| Issue |
Knowl. Manag. Aquat. Ecosyst.
Number 422, 2021
Topical Issue on Fish Ecology
|
|
|---|---|---|
| Article Number | 13 | |
| Number of page(s) | 13 | |
| DOI | https://doi.org/10.1051/kmae/2021011 | |
| Published online | 25 March 2021 | |
Research Paper
First look into the evolutionary history, phylogeographic and population genetic structure of the Danube barbel in Croatia
Premier regard sur l'histoire de l'évolution, la phylogéographie et la structure génétique des populations de barbeau du Danube en Croatie
1
Department of Biology, Faculty of Science, University of Zagreb, Rooseveltov trg 6, Zagreb 10000, Croatia
2
Faculty of Teacher Education, University of Zagreb, Savska cesta 77, Zagreb 10000, Croatia
3
Institute for Environment and Nature, Ministry of Economy and Sustainable Development, Radnička cesta 80, Zagreb 10000, Croatia
* Corresponding author: lucija.raguz@biol.pmf.hr
Received:
19
November
2020
Accepted:
17
February
2021
The Danube barbel, Barbus balcanicus is small rheophilic freshwater fish, belonging to the genus Barbus which includes 23 species native to Europe. In Croatian watercourses, three members of the genus Barbus are found, B. balcanicus, B. barbus and B. plebejus, each occupying a specific ecological niche. This study examined cytochrome b (cyt b), a common genetic marker used to describe the structure and origin of fish populations to perform a phylogenetic reconstruction of the Danube barbel. Two methods of phylogenetic inference were used: maximum parsimony (MP) and maximum likelihood (ML), which yielded well supported trees of similar topology. The Median joining network (MJ) was generated and corroborated to show the divergence of three lineages of Barbus balcanicus on the Balkan Peninsula: Croatian, Serbian and Macedonian lineages that separated at the beginning of the Pleistocene. Croatian lineage of B. balcanicus shows a uniform structure with many recently diverged haplotypes, which was further supported by the star-like shape of the MJ network. Effective size estimates were obtained for populations inhabiting separate river catchments and the results imply the good reproductive potential of Barbus balcanicus in Croatia, which is in positive correlation with the overall high genetic diversity calculated for its populations. Nevertheless, population sizes of several population seem to be reduced as a consequence of habitat degradation and fragmentation.
Résumé
Le barbeau du Danube, Barbus balcanicus, est un petit poisson d'eau douce rhéophile, appartenant au genre Barbus qui comprend 23 espèces indigènes d'Europe. Dans les cours d'eau croates, on trouve trois membres du genre Barbus, B. balcanicus, B. barbus et B. plebejus, chacun occupant une niche écologique spécifique. Cette étude a examiné le cytochrome b (cyt b), un marqueur génétique commun utilisé pour décrire la structure et l'origine des populations de poissons afin d'effectuer une reconstruction phylogénétique du barbeau du Danube. Deux méthodes d'inférence phylogénétique ont été utilisées: la parcimonie maximale (MP) et la probabilité maximale (ML), qui ont permis d'obtenir des arbres bien étayés et de topologie similaire. Le réseau d'assemblage médian (MJ) a été généré et validé pour montrer la divergence de trois lignées de Barbus balcanicus dans la péninsule balkanique: les lignées croate, serbe et macédonienne qui se sont séparées au début du Pléistocène. La lignée croate de B. balcanicus présente une structure uniforme avec de nombreux haplotypes récemment divergents, qui a été renforcée par la forme étoilée du réseau MJ. Des estimations de taille effective ont été obtenues pour des populations vivant dans des bassins hydrographiques séparés et les résultats impliquent le bon potentiel reproductif de Barbus balcanicus en Croatie, qui est en corrélation positive avec la diversité génétique globale élevée calculée pour ses populations. Néanmoins, la taille de plusieurs populations semble être réduite en raison de la dégradation et de la fragmentation de l'habitat.
Key words: Barbels / Barbus balcanicus / phylogenetics / cytochrome b
Mots clés : Barbeaux / Barbus balcanicus / phylogénétique / cytochrome b
© L. Raguž et al., Published by EDP Sciences 2021
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License CC-BY-ND (https://creativecommons.org/licenses/by-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. If you remix, transform, or build upon the material, you may not distribute the modified material.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License CC-BY-ND (https://creativecommons.org/licenses/by-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. If you remix, transform, or build upon the material, you may not distribute the modified material.
1 Introduction
European barbels originated from an East Asian ancestor, migrating to Europe via the Siberian route duringthe Lower Miocene–Upper Oligocene (Tsigenopoulos and Berrebi, 2000) where their radiation developed in the Mediterranean region (Berrebi and Tsigenopoulos, 2003). Today there are 35 valid species of genus Barbus Cuvier & Cloquet 1816 in the world distributed from Iberian Peninsula in Western Europe to Transcaspian region (the Atrek basin) in the Central Asia (Levin et al., 2019). In European watercourses there are around 22 native barbels (Kottelat & Freyhof, 2007) and most recent description was Barbus biharicus from Sebes-Körös River in Hungary and Romania (Antal et al., 2016).
Croatian watercourses are inhabited by three species of the genus Barbus. Two species inhabit watercourses of the Danube (Black Sea) basin: barbel Barbus barbus (Linnaeus, 1758) inhabiting middle and lower courses of rivers draining into the Danube River, and the Danube barbel, Barbus balcanicus described by Kotlík et al., 2002, found in the mountainous streams and small rivers draining to Sava, Drava and Kupa Rivers. The third species, Italian barbel, Barbus plebejus Bonaparte, 1839, inhabits waters of the Adriatic basin on the Istrian Peninsula, and the Krka and Zrmanja Rivers (Ćaleta et al., 2019).
Barbus balcanicus was distinguished from other Barbus species based on mitochondrial DNA (mtDNA) cytochrome b (cyt b), and this was further supported with morphological characters that differed it from B. petenyi Heckel, 1852, the Romanian barbel, as B. balcanicus was previously identified (Kotlík et al., 2002). Their research revealed that Barbus petenyi is actually a species complex comprised of at least three distinct species (B. petenyi sensu stricto, B. balcanicus and B. carpaticus), which likely diverged during the Miocene (Antal et al., 2016). Due to cyclical glaciation and ice retreat periods at the end of Pliocene and throughout the Pleistocene, water connections between catchments were disabled, leading consequently to the isolation of freshwater populations and species separation (Marková et al., 2010).
The Danube barbel is a small rheophilic, lithophilic and benthopelagic freshwater fish that dwells in fast and moderately fast streams with a gravel bottom, mostly residing in riffles and waterfalls. Its distribution area, besides Croatia, includes Bosnia and Herzegovina, Serbia, Macedonia, Southwestern part of Romania, Bulgaria, watercourses of Aegean basin and Isonzo (Soča) River basin (Kottelat & Freyhof, 2007; Buonerba et al., 2015; Bertoli et al., 2019). During the spawning season (May to July), the Danube barbels migrate upstream in search of highly oxygenated currents to spawn (Kottelat & Freyhof, 2007; Žutinić et al., 2014). They are primarily vagile species with high dispersal ability and gene flow expected to occur throughout a river catchment, while migrations between river drainages are likely limited.
Within the family Cyprinidae, species of the genus Barbus have been broadly used for studies of regional biogeography patterns and changes in freshwaters due to their wide distribution, diversity and varied ecological adaptions (Zaccara et al., 2019). Barbels have been the focus of many molecular studies investigating the relationships between species and populations in specific European regions (Tsigenopoulos and Berrebi, 2000). Such studies including Barbus balcanicus have been carried out in Italy (Meraner et al., 2013; Buonerba et al., 2015; Bertoli et al., 2019) Serbia (Simonović et al., 2018), Albania (Marková et al., 2010) and Macedonia (Velkova-Jordanoska et al., 2013), but to date not in Croatia.
Native Italian barbels are considered to be B. balcanicus, B. caninus Bonaparte, 1839, B. plebejus and B. tyberinus Bonaparte, 1839 (Buonerba et al., 2015). Barbus balcanicus primarily colonizes the small tributaries of the Isonzo (Soča) River system near the border with Slovenia, though its distribution in Italy could be wider since it was often erroneously classified as B. caninus given the high morphological similarity. However, Bertoli et al. (2019) stated that ecological preferences of these two species differ, as B. caninus requires oxygenated waters and high current speed while B. balcanicus prefers lower flow rates, cobble/flat rock substrates and less cover vegetation. Populations of B. balcanicus have been found in sympatry with B. plebejus (Pneuma Creek, Piumizza and Reca Streams), indicating possible hybridization (Bertoli et al., 2019).
Recent study of Simonović et al. (2018) revealed that Serbian watercourses are inhabited by B. strumicae Karaman, 1955 (Struma River catchment) and B. balcanicus (Vrla, Krupaja and Gradac River). The Albanian endemic species, B. prespensis Karaman, 1924, believed to exclusively inhabit the Prespa Lakes was later found in the surrounding catchments (Devoll River), and B. rebeli Koller, 1926 is most likely a species complex found to be endemic to several river drainages in western Albania, where it evolved through allopatry in systems of historically interconnected lakes and their basins (Marková et al., 2010).
According to the nuclear gene (a beta-actin intron, Actb) in the Drini River, part of the Ohrid–Drini–Shkodra River system, B. rebeli and B. balcanicus came into secondary contact and hybridized (Marková et al., 2010). They also concluded that B. rebeli from the Pavllo River in Albania is actually B. peloponnesius Valenciennes 1842, thought to be endemic to Greece but its northern limit appears to extend into southern Albania. Velkova-Jordanoska et al. (2013) used Random Amplification of Polymorphic DNA (RAPD-PCR) for determination of Barbus species coexisting in North Macedonia. Barbus macedonicus Karaman, 1928 inhabiting the Vardar and Crna Rivers showed the greatest difference from all other analysed species, with populations differing between Lake Ohrid and the Vardar River, likely representing the still undescribed species Barbus sp. Ohrid-Drini-Shkodra and Barbus balcanicus (Marková et al., 2010).
Throughout the application of mitochondrial marker, cytochrome b (cyt b) this study aimed to fill the gaps regarding the phylogeography, evolutionary history and population genetics of the Danube barbel in Croatia. The genetic marker cytochrome b was selected due to its suitability for taxonomic and population genetic studies as proven in many studies on a wide range of vertebrates (e.g. Tobe et al., 2010; Tominaga et al., 2010; Sagonas et al., 2014). It is considered to be one of the most suitable genes for revealing phylogenetic relationships between closely related taxa (Patwardhan et al., 2014). Cyt b is frequently used for describing the structure and origin of fish populations (e.g. Perea et al., 2010; Buj et al., 2014, 2019, 2020; Schenekar et al., 2014), including molecular studies of the genus Barbus (Tsigenopoulos and Berrebi, 2000; Kotlík et al., 2002; Bertoli et al., 2019; Zaccara et al., 2019).
2 Material and methods
2.1 Sampling and site description
During 2018, 115 samples from 30 populations of the Danube barbel were collected throughout its distribution range in Croatia (Fig. 1, Tab. 1). Sampling was by electrofishing from water using a Hans Grassl backpack electrofishing generator (model IG-200-1) in mountain and highland streams, at elevations from 300 to 1000 m, with moderate climate conditions. For all specimens, a small clip was taken from the right pelvic fin and stored in 96% ethanol, and all fish were safely returned to the water. Due to the broad population dispersion, geographic distance and geological differences, while sampling populations were grouped into subregions corresponding to three Croatian regions: North, Central and East. All sampled localities belong to larger river catchments which further drain into Drava or Sava Rivers which belong to Danube (Black Sea) basin.
In the North region, situated in northwest Croatia around the capital city of Zagreb, 10 populations of Danube barbel were sampled in four subregions: Medvednica, ZelinskoGorje, Ivančica and Sutla. The largest subregion is the Medvednica Nature Park on Mt. Medvednica (1035 m), composed of magmatic, sedimentary and metamorphic rock and originating in the Pliocene and later the Quaternary (Brkić, 2017). Although covered with rich vegetation, numerous karst forms are present on Mt. Medvednica, including caves, pits, karst valleys and sinkholes. The second subregion, the ZelinskoGorje plateau (330 m), lies to the north of Mt. Medvednica. The subregion Mt. Ivančica (1061 m) is at northernmost point of Danube barbel's distribution in Croatia and to the west is the Sutla River, considered a small, distinctive subregion. The watercourses of Zagorje drain into two separate drainages, the Drava and Sava Rivers. The Željeznica and Žarovnica Streams feed the Bednja River, a tributary of the Drava River. The Lonjica, Kašina and Burnjak Streams feed the Lonja River, a tributary of the Sava River. The streams of Mt. Medvednica (Sarni, Pronjak and Rijeka) drain into the Krapina River, a tributary to the Sava River. The Vidovec Stream downstream merges with lowland streams of Zagreb and through a channel connects to Sava River.
The most populations (15) were found in the East region, Slavonia. Four subregions were identified in this region, each associated with a separate hilly or mountainous area: Mt. Papuk (953 m), a nature park due to its geological features and forest wilderness, Mt. Krndija (792 m) to the east, Ravna Gora hill (854 m) to the west, and Požeška Gora hill (618 m) to the south. Each of these features lifted during the Pliocene and Quaternary (Brkić, 2017). The waters from these hills and mountains primarily drain into the Pakra River, which feeds the Lonja River, a tributary to the Sava River. Česma River, also drains into Lonja River which further connects to Sava River. The only Slavonian populations belonging to the Drava River drainage are Velika Radetina and Šumečica, while the Orljava and Rešetarnica Rivers are both the tributaries to the Sava River.
In the Central region five populations were sampled in two subregions: Banija (with the Zrinska Gora hills (peak at 615 m)) and the Petrova Gora hills (peak at 512 m). Both are composed of rocks of Mesozoic origin, nested between two river drainages, the Kupa and Sava Rivers. Most streams in this region belong to the Glina River catchment, a tributary to the Kupa River, with the exception of the Stupnica Stream and Sunja River. The Stupnica Stream flows into the Una River that springs in Croatia, flows through a corner of Bosnia-Herzegovina before draining into the Sava River. The Sunja River is a direct tributary of the Sava River.
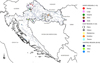 |
Fig. 1 Map of sample localities of the Danube barbel in Croatia. Each colour corresponds to a river catchment. |
Distribution of cytochrome b haplotypes of the Danube barbel in the populations, river catchments, river drainages and regions of its Croatian distribution range.
3 Laboratory work
Total genomic DNA was isolated from fin tissue samples using the DNeasy Blood and Tissue isolation Kit (Qiagen), following the manufacturer's instructions. Polymerase chain reactions (PCR) were prepared using Hot Star Taq Plus Master Mix Kit (Qiagen) and a specific set of primers, Glu-F 5' GAAGAACCACCGTTGTTATTCAA 3' and Thr-R 5' ACCTCCRATCTYCGGATTACA 3' (Zardoya and Doadrio, 1998), used to amplify the cytochrome b gene in its full length, 1141 base pairs (bp). The prepared PCR reactions contained 12.5 μL Hot Star Taq Plus Master Mix (Qiagen), 2.5 μL CoralLoad Concentrate (Qiagen), 2 μL RNase-Free Water, 2 μL of each primer and 4 μL isolated DNA, in total 25 μL. The PCR protocol was optimized as following: first denaturation step, 10 min at 95 °C; 35 cycles − denaturation for 45 s at 92 °C, annealing for 90 s at 48 °C, extension for 105 s at 72 °C and the final polymerisation step for 7 min at 72 °C. Sequencing was conducted by Macrogen Europe (Netherlands). For sequence alignment, BioEdit 7.2.5 software (Hall, 1999) was used, and all chromatograms were visually checked while aligning.
3.1 Data analysis
Phylogenetic reconstruction was based on two methods of phylogenetic inference: maximum parsimony (MP) and maximum likelihood (ML), using PAUP 4.0b10 (Swofford, 2002). For MP analysis, the heuristic search mode was used with randomized input orders of taxa, and TBR branch swapping with all codon sites and nucleotide substitutions types weighted equally. Nonparametric bootstrapping (1000 pseudo-replicates for MP, 100 for ML, 10 additional sequence replicates) was used to assess branch support (BS). Alongside haplotypes from Croatia, haplotypes of other Barbus species available in the full length of the cytb gene were retrieved from GenBank (Tab. 2) and included in the phylogenetic reconstruction. For tree rooting, we used a sequence of Cyprinus carpio Linneaus, 1758. The median joining (MJ) network was created using Network 10.0.0.0. (Fluxus Technology Ltd.), as phylogenetic networks are useful for untangling the relationships between closely related taxa as they do not exclude recombination, hybridization and horizontal gene transfer (Posada and Crandall, 2001). Only the haplotypes of Barbus balcanicus, from this study and from GenBank, were included in the MJ network.
To check whether all mutations were selectively neutral, statistical tests D and F (Fu and Li, 1993) and Tajima's test (Tajima, 1989) were conducted on data sets using DnaSP v5 (Librado and Rozas, 2009). The level of genetic diversity within each population of Danube barbel, river catchment and region were described using several measures of DNA polymorphism (haplotype number, haplotype diversity, number of polymorphic sites, total number of mutations, average number of nucleotide differences, nucleotide diversity). These tests were conducted using DnaSP 5.10 (Librado and Rozas, 2009).
Using MIGRATE 3.2.1 software (Beerli and Felsenstein, 2001; Beerli, 2009), effective populations sizes were calculated in each river catchment. Theta values were generated from the FST-calculation, using a migration matrix model with variable theta and constant mutation rate. Analysis strategy was a maximum likelihood, MCMC model with the settings: 10 short chains with 20 000 sampled and 1000 recorded trees and 3 long with 200 000 sampled and 10 000 recorded trees, 2 averaging over replicates. Two static heating schemes, 4 chains with temperatures: 1.00, 1.57, 2.71, 5.00; swapping interval of 1, and 10 000 discard trees per chain. Calculated thetas (θ) were used to estimate population effective sizes (Ne), using a mutation rate calculated for the Cyprinidae family of 0.76 % per pairwise comparison per million years (MY) (Zardoya and Doadrio, 1999), which was further converted to mutation per sequence per generation, μ = 0.0000023. For Ne, both the MLE (maximum likelihood estimate) and 5–95% levels (estimates at 5th and 95th probability percentile) were estimated.
Estimation of divergence times of distinct evolutionary lineages was based on the cyt b sequences obtained here, with those retrieved from GenBank. Analyses were conducted using the Bayesian MCMC coalescent method in Beast 1.7.0 software (Drummond et al., 2012). A relaxed log normal clock was applied, which assumes that the substitution rates associated with each branch are independently drawn from a single, discretized lognormal distribution (Drummond et al., 2006). For molecular clock calibration we have used previously published data on divergence events (Gante, 2011), as well as on the Barbus fossil evidences (Levin et al., 2019). We have chosen three nodes in our phylogenetic tree that were comparable with previously reported phylogenies (origin of Barbus, origin of B. peloponnesius and the onset of intraspecific divergences inside B. balcanicus) and used data of Gante (2011) and Levin et al. (2019) to calibrate those nodes (18.5 ± 0.5 MYA, 10.8 ± 2.1 MYA and 1.9 ± MYA, respectively). Branch rates were drawn from an uncorrelated lognormal distribution and a speciation birth-death tree prior with random starting tree. The substitution model applied was HKY with Gamma site heterogeneity. Default prior distributions were applied for kappa, frequencies and alpha, whereas substitution rate parameters were unlinked across codon position. The number of MCMC steps (the length of chain) was 10 million and ESS values for all parameters were well above 200 (543–879).
Danube barbel cyt b sequences retrieved from GenBank included in the phylogenetic and evolutionary history reconstruction.
4 Results
4.1 Phylogenetic reconstruction and divergence time estimations
A total of 125 individuals of Barbus balcanius were sampled at 30 localities in Croatia (Tab. 1). We successfully obtained 115 sequences of cytochrome b in the full length of the gene (1141 bp). Within the total sample, 15 new cyt b haplotypes were identified, 4 as private haplotypes present only in a single population (CRO 12–Željeznica, CRO 13–Velika Radetina 1&2, CRO 14–Bijela rijeka, and CRO 15–Brusnica). Neutrality tests suggested no deviation from mutation-drift equilibrium for the data set, D*, F* and Tajima's D but tests showed no statistical significance. No insertions or deletions were observed in the sequence set. In the phylogenetic reconstruction, haplotypes of other European Barbus species were included, for a total of 47 haplotypes, with Cyprinus carpio as the outgroup. Of 1141 bp, there were 359 variable (31.46%) and 261 parsimony informative characters (22.87%). The length of the MP tree was 725, with a consistency index of 0.5683, homoplasy index of 0.4317 and retention index of 0.8554.
The two different phylogenetic reconstruction methods yielded trees of similar topologies (Fig. 2), revealing three clades of Barbus species. The first comprises B. balcanicus, B. carpathicus and B. caninus. All Croatian haplotypes of B. balcanicus clustered together with one haplotype from Bosnia and Hercegovina (BIH) and one from Serbia (SRB 3). Both trees showed that the Croatian haplotypes are closer to Serbian than Macedonian haplotypes, with SRB 3 haplotype as the closest. Obtained trees showed that the closest relative to B. balcanicus is B. carpathicus while B.caninus is a sister taxon to both of them. The second clade included species of the southern Balkans, B. peloponnesius, B. petenyi, B. rebeli and B. prespensis with sister taxon B. sp. ODS, a still undescribed species from Ohrid-Drini-Shkodra system. The third clade included B. macedonicus and B. tyberinus and two other Barbus species that could be found in Croatia, B. barbus and B. plebejus, with B. tyberinus positioned as a sister taxon to B. plebejus. Sequences of the Spanish B. meridionalis were separate from these three clades in the Barbus genus.
In Croatia, the Danube barbel has a uniform structure with many shallowly separated haplotypes, as further supported by the star shape of the MJ network (Fig. 3) centred around CRO 4. The MJ network (Fig. 3) including only haplotypes of Barbus balcanicus (from this study and GenBank) showed that most haplotypes are present in several river catchments. All Croatian haplotypes clustered around CRO 4 which is widely distributed and appears to be ancestral. All other haplotypes originated from this haplotype, and most differ in a single nucleotide. CRO 9 is divergent from CRO 4 due to one mutation, and CRO 10 and 12 emerged from CRO 9; all are found in the Bednja River catchment. Haplotypes from Serbia and Macedonia clustered separately from each other and from Croatian haplotypes, except SRB 3 (B. balcanicus Jadar River, Serbia) which is one mutation apart from CRO 4, same as haplotype BIH (B. balcanicus Vrbas River, Bosnia and Herzegovina). Haplotype SRB 1 (B. balcanicus Džepska River 1, Serbia) was 8 mutations apart from CRO 4, and MKD 1 (B. balcanicus Lepenac River 1, Macedonia) was 11 mutations apart.
Analyses of the evolutionary history using Beast software package (Drummond et al., 2012) revealed that the genus Barbus has an ancient origin, separating from the genus Cyprinus around 41 (26.07–56.51) million years ago (MYA), during the Middle Eocene (Fig. 4). The first split of Barbus species happened around 18.43 (17.47–19.41) MYA in the Burdigalian age of the Early Miocene, when two clades diverged, B. carpathicus and B. balcanicus and separated from other Barbus species. The Danube barbel separated from its sister taxon B. carpathicus during the Late Miocene in the Tortonian age. During the Pliocene, Paratethys was divided into several inland seas, including the Pannonian Sea, which dried up in the early Pleistocene (Kázmér, 1990) when subsequent diversification events occurred, starting with the first glaciation events in the Gelasian age. The Croatian lineage of B. balcanicus is of younger origin and its interlineage divarication occurred around 0.68 (0.38–1.04) MYA, corresponding with the Günz glaciations.
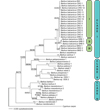 |
Fig. 2 ML phylogram based on cyt b sequences (1141 bp long) showing the position of Danube barbel populations in Croatia within the genus Barbus. Numbers at nodes represent MP and ML bootstrap values; an asterisk replaces bootstrap values of 100. |
 |
Fig. 3 MJ network obtained from cyt b sequences (1141 bp) of Barbus balcanicus. showing the star-shaped phylogenetic distribution of Croatian haplotypes. Circle size is proportionate to haplotype frequencies and each colour represents a river catchment. Dashed lines represent mutations. |
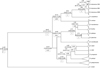 |
Fig. 4 Divergence time estimations based on cyt b squences of Barbus species. Timing of splitting events is presented as the mean value and 95% credibility range (in million years ago). The lower bound and upper bound of the highest posterior density (HPD) interval, the HPD is the shortest interval that contains 95% of the sampled values. Numbers in squares represent bootstrap values. |
4.2 Genetic diversity and effective size estimations of Danube barbel populations
The calculated DNA polymorphism measures provide insight into the genetic structure and diversity of populations, river catchments and regions of the Barbus balcanicus distribution range in Croatia (Tab. 3). Six populations had only one haplotype, which may be due to the small sample size from those localities (≤4). Populations showing lower genetic diversity were Vidovec and Brusovača, where the calculated haplotype diversity (Hd) for those localities was ≤0.4 and π of 0.00029 (Vidovec) to 0.00070 (Brusovača). The Kašina population had the highest number of haplotypes, with 5 haplotypes in just 8 individuals. The highest genetic diversity was calculated for Lonja River catchment, within 20 individuals there were 8 haplotypes and Hd of 0.884 supported by π of 0.00135. Followed by Bednja River catchment with 4 haplotypes within 8 individuals, Hd of 0.821 and π of 0.00166. River catchments that comprised more than one population (Krapina, Pakra, Orljava, Karašica and Glina) all had 4–5 haplotypes within 10–15 sampled individuals. Haplotype diversity, Hd for those river catchments varies from 0.733 (Orljava) to 0.781 (Krapina) and nucleotide diversity, π varies from 0.00090 (Karašica) to 0.00117 (Krapina). Higher diversity was recorded in two river catchments that were represented with only one population, Rešetarnica and Sunja, both had Hd of 0.8. Although, Sunja River catchment had higher π of 0.00158 since 5 individuals carried 3 haplotypes and for Rešetarnica River catchment, 4 haplotypes were found inside 10 individuals so their nucleotide diversity, π was estimated to 0.00099. For other river catchments comprising only one population very low genetic diversity was calculated, most with just one haplotype, only Sava River catchment (not the whole Sava River drainage) represented with population of Vidovec Stream had two haplotypes, Hd of 0.33 and π 0.00029. At the regional scale, high genetic diversity was noted for all three regions: North region had a Hd = 0.883 with 10 haplotypes and π of 0.00140, East region had a Hd = 0.804 with 11 haplotypes and π of 0.00102, and Central region with the highest estimated population variability with Hd = 0.869, 7 haplotypes inside 18 individuals and π of 0.00150. Understanding the gene diversity of each population is important for estimating its ability to cope with environmental changes.
Effective size estimates were conducted for each river catchment (Tab. 4), both the MLE (maximum likelihood estimate) and 5–95% levels (estimates at 5th and 95th probability percentile). The results suggest the good reproductive potential of Barbus balcanicus in Croatia and high probability of its survival, although calculations did not show positive correlations for all catchments. The highest number of individuals was estimated in the East region, in the Drava and Pakra catchments (Ne (MLE) = 1043 and 652, and Ne (5–95%) = 478-* and 430–1043, respectively). The fewest individuals in the East region was calculated for the Orljava River, despite containing four populations, while the Rešetarnica River catchment had lower Ne 113 (82–152) with only the Putnjak population in its tributary. In the North region, the Bednja and Lonja River catchments showed equal estimations (Ne (MLE) = 217), while the Krapina and Sava River catchments showed lower estimates (Ne (MLE) = 61 and 78). In the smallest Central region, the Glina River catchment and the catchments Brusovača, Listovački stream and Bublen, showed very low Ne (MLE) of only 65, while the Sunja River catchment, as an isolated watercourse without tributaries, had a Ne (MLE) of 234.
Genetic polymorphism measures based on the cyt b gene for Barbus balcanicus in Croatia.
Effective size estimates for river catchments.
5 Discussion
5.1 Phylogenetic position of Barbus balcanicus in Croatia within the genus Barbus and its evolutionary history
This study revealed that the Danube barbel in Croatia, based on mitochondrial DNA, has a uniform structure with many shallowly separated haplotypes, indicating recent colonization from a single glacial refugium. This origin theory was further supported with the MJ phylogenetic network and its star shape, centred around the CRO 4 haplotype as the ancestral one. Nevertheless, the Croatian haplotypes clustered separately from three Serbian and three Macedonian haplotypes, revealing that there are at least three intraspecific lineages of Barbus balcanicus on the Balkan Peninsula. One Serbian haplotype and one from Bosnia and Herzegovina clustered with Croatian haplotypes. The Danube barbel separated from other Barbus species during the Miocene (Marková et al., 2010; Berrebi et al., 2014; Antal et al., 2016). This includes its sister taxon Barbuspetenyi (Kotlík et al., 2002) from which it diverged around 18.43 MYA. Supported by its distant position in the obtained trees, B. petenyi appears closest to B. rebeli and B. prespensis. The Danube barbel diverged later from its sister taxon Barbus carpathicus (Kotlík et al., 2002), 8.54 MYA in the Late Miocene, and their position was the same on both phylogenetic and divergence time trees.
The haplotype CRO 4 is most widely distributed in Croatia and found throughout the distribution range of this species, implying the possibility that the first colonizers after glaciation bore this haplotype and the remaining ones diverged from it later. One haplotype from Bosnia and Herzegovina (BIH) and one from Serbia (SRB 3) also trace their origin to CRO 4. Furthermore, Croatian lineage separated from Macedonian lineage around 1.88 MYA and from Serbian (without haplotype SRB 3) 1.26 MYA.Both splits correspond with glaciation and ice retreat periods that alternated throughout the Pleistocene, resulting in the isolation of freshwater populations and species separation (Marková et al., 2010).
Analysing the MP and ML tree, we can conclude that Barbus balcanicus in the Balkan Peninsula is monophyletic. Throughout its Croatian distribution range, the Danube barbel adapted to the specific characteristic of watercourses and habitats in different regions, as seen in the genetic differences and multitude of haplotypes. Species of genus Barbus are considered to be tetraploid (Berrebi, 1995) and polyploids are considered to have greater tolerance to ecological variation due to the duplication of their genes, which provides metabolic flexibility (Tsigenopoulos et al., 2002). Tetraploidy could have induced Barbus balcanicus dispersion around the Balkan Peninsula and helped it to cope with environmental challenges.
The overall position of sequences retrieved from GenBank follows previously published papers (Marková et al., 2010; Antal et al., 2016; Bertoli et al., 2019). The Italian B. caninus distributed in northern Italy and Slovenia in the tributaries of upper Adriatic basin, from the Isonzo (Soča) River in the north to the Marecchia River in the south (Bianco and Delmastro, 2004) and B. carpathicus which inhabits Czech Republic, Hungary, Poland, Romania, Slovakia and Ukraine appear to be the closest relatives to B. balcanicus.
5.2 Population genetic estimates of Barbus balcanicus in Croatia
The Danube Barbel in Croatia shows relatively high genetic polymorphism, indicated by the high genetic diversity (overall Hd = 0.88 and π = 0.00130), despite the relatively low number of haplotypes (15 haplotypes in a sample of 115 sequences). When compared to other Cypriniform species in Croatia, the observed values of genetic diversity in this species is significantly lower than polymorphisms of species distributed in karst rivers, despite their more restricted distribution ranges (e.g., Telestes polylepis Steindachner, 1866 distributed in a single lake has a Hd of 0.9; T. tursky (Heckel, 1843) in a single small river has a Hd of 0.964 (Buj et al., 2017); Cobitis dalmatina Karaman, 1928 distributed in the Cetina River has a Hd of 0.99 (Buj et al., 2014); all the above species have much narrower distribution ranges than B. balcanicus in Croatia). The reason for this difference is the very old origin and unconstrained evolutionary histories of these karst species. On the other hand, the Croatian lineage of B. balcanicus is of much younger origin, diverging around 0.68 MYA, despite the much older origin of the species. Its evolutionary development during this period resulted in the observed level of genetic diversity. Few species in continental Croatia have been examined for their genetic diversity to date. Moreover, there is a lack of data on species with similar ecological requirements as B. balcanicus, inhabiting upper parts of rivers and streams and living in colder, well oxygenated waters. Salmo labrax Pallas, 1814 in the Plitvice Lakes system expressed moderate genetic diversity, estimated using cytochrome b as well, Hd = 0.615 (Buj et al., 2020), whereas its diversity within the Žumberak-SamoborskoGorje Nature Park Hd = 0.807 (Ivić et al., 2020) is similar to the genetic polymorphism of B. balcanicus in Croatia.
Though the observed genetic diversity in several populations was zero (at the localities Hromec, Listovački stream, Sivornica, Račačka rijeka, Kutjevačka rijeka and Rinovica) or very low (Hd = 0.33 in the Vidovec stream), these estimates may not be reliable due to the small sample size. Genetic diversities observed in populations with an adequate sample size were moderate to high and the observed genetic polymorphism of B. balcanicus is likely due to its wide distribution in rivers and streams of the Danube basin in Croatia, and also due to some 680 000 years of independent evolution. With the exception of the Karašica and Pakra River catchments, populations of the Danube barbel in Croatia have low effective population sizes (61–234), which might be a consequence of the smaller stretches of adequate habitats in individual rivers and streams. However, this may indicate that some populations have reduced sizes due to anthropogenic pressures. Unfortunately, majority of localities included in this investigation are not in the natural or near-natural state. Habitat fragmentation and degradation are present on many water bodies inhabited by B. balcanicus and our result indicate that they are very problematic for its populations. Namely, by far the highest effective population size was estimated for population inhabiting the only localities that are still in natural state, without any habitat degradation or other threats − Velika Radetina and Šumečica streams in the Karašica River catchment. On the other localities, where habitat degradation is present, sometimes even coupled with pollution (such as are localities in the Bednja, Lonja and Pakra River catchments), effective population sizes seem to be reduced (estimated to 217–652 individuals). The lowest densities are characteristic for populations in rivers and streams affected by habitat fragmentation (such as localities in the Krapina River catchment, where effective population size is estimated to only 71 individuals), and localities where habitat degradation and pollution are particularly intense (for example the Vidovec stream, which is completely channelized and very polluted, and the effective population size of the Vidovec population is only 61). Noteworthy, besides on the highly affected habitats, small population sizes were observed also on the localities in the Glina River catchment, which have not been anthropogenically modified. However, those are mostly very small streams that, at least partially, dry out during part of the year, so it is likely that they only present temporary habitat for this species. Generally, we can conclude that habitat fragmentation, particularly if coupled with habitat degradation and pollution, is the most problematic threat to B. balcanicus, leading to reductions in its population sizes. This conclusion is concordant with life history traits of B. balcanicus, because it migrates in the upper parts of rivers and streams for reproduction (Kottelat & Freyhof, 2007), but also previous reports (Mrakovčić et al., 2006). Thereafter, habitat protection and restoration, particularly removal of barriers and/or mitigation of negative effects of habitat fragmentation should make up the most important parts of conservation plans.
Acknowledgments
The authors are grateful to SinišaVajdić and Roman Karlović for their dedicated help during the fish sampling and to Linda Zanella for proofreading the paper.
References
- Antal L, László B, Kotlík P, Mozsár A, Czeglédi I, Oldal M, Kemenesi G, Jakab F, Nagy SA. 2016. Phylogenetic evidence for a new species of Barbus in the Danube River basin. Mol Phylogenet Evol 96: 187–194. [Google Scholar]
- Beerli P. 2009. How to use Migrate or why are Markov Chain Monte Carlo programs difficult to use. In Bertorelle G, Bruford MW, Hauffe HC, Rizzoli A, Venersi C, eds. Population Genetics for Animal Conservation, Vol. 17. Cambridge: Cambridge University Press, pp. 42– 79. [Google Scholar]
- Beerli P, Felsenstein J. 2001. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Genetics 152: 763–773. [Google Scholar]
- Benovics M, Vukić J, Šanda R, Rahmouni I, Šimková A. 2020. Disentangling the evolutionary history of peri-Mediterranean cyprinids using host-specific gill monogeneans. Int J Parasitol 50: 969–984. [Google Scholar]
- Berrebi P. 1995. Speciation of the genus Barbus in the North Mediterranean basin: recent advances from biochemical genetics. Biol Conserv 72: 237–249. [Google Scholar]
- Berrebi P, Tsigenopoulos C. 2003. Phylogenetic organisation of the genus Barbus sensu stricto − A review based on data obtained using molecular markers. In: Bǎnǎrescu PM, Bogutskaya NG, Eds. Freshwater fishes of Europe − Cyprinidae 2, Part II − Barbus, Vol. 5 II. AULA-Verlag GmbH, Wiebelsheim, pp. 11– 22. [Google Scholar]
- Berrebi P, Chenuil A, Kotlík P, Machordom A, Tsigenopoulos C. 2014. Disentangling the evolutionary history of the genus Barbus sensu lato, a twenty years adventure. In: Professor Carlos Almaça (1934–2010) Maria Judite Alves, Alexandra Cartaxana, Alexandra Marçal Correia and Luis Filipe Lopes, eds. Museu Nacional de História Natural e da Ciência, Museu Nacional de História Natural e da Ciência Universidade de Lisboa. Rua da Escola Politécnica 56/58, 1250–102 Lisboa (Portugal), pp. 29– 55. [Google Scholar]
- Bertoli M, Pizzul E, Devescovi V, Franz F, Pastorino P, Giulianini PG, Ferrari C, Nonnis Marzano F. 2019. Biology and distribution of Danube barbel (Barbus balcanicus) (Osteichthyes: Cyprinidae) at the Northwestern limit of its range. Eur Zool J 86: 280–293. [Google Scholar]
- Bianco PG, Delmastro G. 2004. Threatened fishes of the world: Barbus caninus Bonaparte, 1839 (Cyprinidae). Environ Biol Fish 71: 352. [Google Scholar]
- Bianco PG, Ketmaier V. 2005. Anthropogenic changes in the freshwater fish fauna of Italy, with reference to the central region and Barbus graellsii, a newly established alien species of Iberian origin. J Fish Biol 59: 190–208. [Google Scholar]
- Brkić Ž. 2017. The relationship of the geological framework to the Quaternary aquifer system in the Sava River valley (Croatia). J Croat Geol Surv Croat Geol Soc 70: 201–213. [Google Scholar]
- Buj I, Šanda R, Marčić Z, Ćaleta M, Mrakovčić M. 2014. Combining morphology and genetics in resolving taxonomy − a systematic revision of spined loaches (Genus Cobitis; Cypriniformes, Actinopterygii) in the adriatic watershed. PLoS ONE 9: e99833. [Google Scholar]
- Buj I, Marčić Z, Ćaleta M, Šanda R, Geiger MF, Freyhof J, Machordom A, Vukić J. 2017. Ancient connections among the European rivers and watersheds revealed from the evolutionary history of the genus Telestes (Actinopterygii; Cypriniformes). PLoS ONE 12: e0187366. [Google Scholar]
- Buj I, Marčić Z, Čavlović K, Ćaleta M, Tutman P, Zanella D, Duplić A, Raguž L, Ivić L, Horvatić S, Mustafić P. 2019. Multilocus phylogenetic analysis helps to untangle the taxonomic puzzle of chubs (genus Squalius: Cypriniformes: Actinopteri) in the Adriatic basin of Croatia and Bosnia and Herzegovina. Zool J Linn Soc 20: 1–22. [Google Scholar]
- Buj I, Raguž L, Marčić Z, Ćaleta M, Duplić A, Zanella D, Mustafić P, Ivić L, Horvatić S, Karlović R. 2020. Plitvice Lakes National park harbors ancient, yet endangered diversity of trout (genus Salmo). J Appl Ichthyol https://doi.org/10.1111/jai.14120 [Google Scholar]
- Buonerba L, Zaccara S, Delmastro GB, Lorenzoni M, Salzburger W, Gante GF. 2015. Intrinsic and extrinsic factors act at different spatial and temporal scales to shape population structure, distribution and speciation in Italian Barbus (Osteichthyes: Cyprinidae). Mol Phylogenet Evol 89: 115–129. [Google Scholar]
- Ćaleta M, Marčić Z, Buj I, Zanella D, Mustafić P, Duplić A, Horvatić S. 2019. A review of extant Croatian freshwater fish and lampreys − annotated list and distribution. Croat J Fish 77: 137–234. [Google Scholar]
- Doadrio I, Carmona JA, Machordom A. 2002. Haplotype diversity and phylogenetic relationships among the Iberian barbels (Barbus, Cyprinidae) reveal two evolutionary lineages. J Hered 93: 140–147. [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol 4: e88. [CrossRef] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [CrossRef] [PubMed] [Google Scholar]
- Fu YX, Li WH. 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [PubMed] [Google Scholar]
- Gante HF. 2011. Diversification of Circum-Meditteranean Barbels, Changing Diversity in Changing Environment, In: Grillo O, ed. InTech, Available from: http://www.intechopen.com/books/changing-diversity-in-changing-environment/diversification-of-circum-mediterranean-barbels , 283–298. [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- Ivić L, Buj I, Raguž L, Marčić Z, Ćaleta M, Zanella D, Mustafić P, Horvatić S. 2020. Diversity and structure of trout populations (Salmo sp., Salmonidae, Actinopteri) in the Žumberak-SamoborskoGorje Nature Park in Croatia. Fundam Appl Limnol https://doi.org/10.1127/fal/2020/1283 [Google Scholar]
- Kázmér M. 1990. Birth, life and death of the Pannonian Lake. Palaeogeogr. Palaeoclimatol Palaeoecol 79: 171–188. [Google Scholar]
- Kotlík P, Tsigenopoulos S, Rab P, Berrebi P. 2002. Two new Barbus species from the Danube River basin, with redescription of Barbus petenyi (Teleostei: Cyprinidae). Folia Zool 51: 227–240. [Google Scholar]
- Kottelat M, Freyhof J. 2007. Handbook of European freshwater fishes. Kottelat, Cornol, Switzerland and Freyhof, Berlin, Germany. [Google Scholar]
- Levin BA, Gandlin AA, Simonov ES, Levina MA, Barmintseva AE, Japoshvili B, Mugue NS, Mumladze L, Mustafayev NJ, Pashkov AN, Roubenyan HR, Shaplovalov MI, Doadrio I. 2019. Phylogeny, phylogeography and hybridization of Caucasian barbels of the genus Barbus (Actinopterygii, Cyprinidae). Mol Phylogenet Evol 135: 31–44. [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [CrossRef] [PubMed] [Google Scholar]
- Machordom A, Doadrio I. 2001. Evolutionary history and speciation modes in the cyprinid genus Barbus . Proc Royal Soc B 268: 1297–1306. [Google Scholar]
- Marková S, Sanda R, Crivelli A, Shumka S, Wilson IF, Vukic J. 2010. Nuclear and mitochondrial DNA sequence data reveal the evolutionary historyof Barbus (Cyprinidae) in the ancient lake systems of the Balkans. Mol Phylogenet Evol 55: 488–500. [Google Scholar]
- Meraner A, Venturi A Ficetola GF, Rossi A, Candiotto A, Gandolfi A. 2013. Massive invasion of exotic Barbus barbus and introgressive hybridization with endemic Barbus plebejus in Northern Italy: where, how and why? Mol Ecol 22: 5295–5312. [PubMed] [Google Scholar]
- Mrakovčić M, Brigić A, Buj I, Ćaleta M, Mustafić P, Zanella D. 2006. Red Book of Freshwater Fish of Croatia. Ministry of Culture, State Institute for Nature Protection, Republic of Croatia, Zagreb. [Google Scholar]
- Patwardhan A, Ray S, Roy A. 2014. Molecular markers in phylogenetic studies: a review. J Phylogenet Evol Biol 2: 131. [Google Scholar]
- Perea S, Bohme M, Župančič P, Freyhof J, Šanda R, Ozulug M, Abdoli A, Doadrio I. 2010. Phylogenetic relationships and biogeographical patterns in Circum-Mediterranean Subfamily Leuciscinae (Teleostei, Cyprinidae) inferred from both mitochondrial and nuclear data. BMC Evol Biol 10: 265. [CrossRef] [PubMed] [Google Scholar]
- Posada D, Crandall KA. 2001. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol Evol 16: 37–45. [PubMed] [Google Scholar]
- Sagonas K, Poulakakis N, Lymberakis P, Parmakelis A, Pafilis P, Valakosa ED. 2014. Molecular systematics and historical biogeography of the green lizards (Lacerta) in Greece: Insights from mitochondrial and nuclear DNA. Mol Phylogenet Evol 76: 144–154. [Google Scholar]
- Schenekar T, Lerceteau-Köhler E, Weiss S. 2014. Fine-scale phylogeographic contact zone in Austrian brown trout Salmo trutta reveals multiple waves of post-glacial colonization and a pre-dominance of natural versus anthropogenic admixture. Conserv Genet 15: 561–572. [Google Scholar]
- Simonović P, Marić S, Tošić A, ŠkrabaJurlina D, Nikolić V. 2018. Morphological and molecular differentiation between Rheophilic Barbels Barbus spp. (Cyprinidae) from headwaters at the divide between drainage areas of the Danube, Vardar and Struma Rivers, Central Balkans. Acta Zool Bulg 70: 153–164. [Google Scholar]
- Swofford DL. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), Version 4 [Computer software and manual]. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [PubMed] [Google Scholar]
- Tobe SS, Kitchener AC, Linacre AM. 2010. Reconstructing mammalian phylogenies: a detailed comparison of the cytochrome b and cytochrome oxidase subunit I mitochondrial genes. PLoS ONE 5: e14156. [Google Scholar]
- Tominaga A, Hidetoshi O, Matsui M. 2010. Phylogeny and phylogeography of the sword-tailed newt, Cynopsensicauda (Amphibia: Caudata), as revealed by nucleotide sequences of mitochondrial DNA. Mol Phylogenet Evol 54: 910–921. [Google Scholar]
- Tsigenopoulos CS, Berrebi P. 2000. Molecular phylogeny of North Mediterranean freshwater barbs (genus Barbus: Cyprinidae) Inferred from cytochrome b sequences: biogeographic and systematic implications. Mol Phylogenet Evol 14: 165–179. [Google Scholar]
- Tsigenopoulos CS, Rab P, Naran D, Berrebi P. 2002. Multiple origins of polyploidy in the phylogeny of southern African barbs (Cyprinidae) as inferred from mtDNA markers. Heredity 88: 466–473. [PubMed] [Google Scholar]
- Velkova-Jordanoska L, Panov S, Kostov V, Stojanovski S, Kostoski G. 2013. Molecular identification of the four species of the genus Barbus in lakes and rivers of R. Macedonia. Nat Montenegrina 12: 687–699. [Google Scholar]
- Zardoya R, Doadrio I. 1998. Phylogenetic relationships of Iberian cyprinids: systematic and biogeographical implications. Proc Royal Soc Biol Sci 265: 1365–1372. [Google Scholar]
- Zardoya R, Doadrio I. 1999. Molecular evidence on the evolutionary and biogeographical patterns of European Cyprinids. J Mol Evol 49: 227–237. [CrossRef] [PubMed] [Google Scholar]
- Zaccara S, Quadroni S, De Santis V, Vanetti I, Carosi A, Britton R, Lorenzoni M. 2019. Genetic and morphological analyses reveal a complex biogeographic pattern in the endemic barbel populations of the southern Italian peninsula. Ecol Evol 9: 10185–10197. [Google Scholar]
- Žutinić P, Jelić D, Jelić M, Buj I. 2014. A contribution to understanding the ecology of the large spot barbel −sexual dimorphism, growth and population structureof Barbus balcanicus (Actinopterygii; Cyprinidae) in Central Croatia. North-West. J Zool 10: 158–166. [Google Scholar]
Cite this article as: Raguž L, Buj I, Marčić Z, Veble V, Ivić L, Zanella D, Horvatić S, Mustafić P, Ćaleta M, Sabolić M. 2021. First look into the evolutionary history, phylogeographic and population genetic structure of the Danube barbel in Croatia. Knowl. Manag. Aquat. Ecosyst., 422, 13.
All Tables
Distribution of cytochrome b haplotypes of the Danube barbel in the populations, river catchments, river drainages and regions of its Croatian distribution range.
Danube barbel cyt b sequences retrieved from GenBank included in the phylogenetic and evolutionary history reconstruction.
Genetic polymorphism measures based on the cyt b gene for Barbus balcanicus in Croatia.
All Figures
 |
Fig. 1 Map of sample localities of the Danube barbel in Croatia. Each colour corresponds to a river catchment. |
| In the text | |
 |
Fig. 2 ML phylogram based on cyt b sequences (1141 bp long) showing the position of Danube barbel populations in Croatia within the genus Barbus. Numbers at nodes represent MP and ML bootstrap values; an asterisk replaces bootstrap values of 100. |
| In the text | |
 |
Fig. 3 MJ network obtained from cyt b sequences (1141 bp) of Barbus balcanicus. showing the star-shaped phylogenetic distribution of Croatian haplotypes. Circle size is proportionate to haplotype frequencies and each colour represents a river catchment. Dashed lines represent mutations. |
| In the text | |
 |
Fig. 4 Divergence time estimations based on cyt b squences of Barbus species. Timing of splitting events is presented as the mean value and 95% credibility range (in million years ago). The lower bound and upper bound of the highest posterior density (HPD) interval, the HPD is the shortest interval that contains 95% of the sampled values. Numbers in squares represent bootstrap values. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


