| Issue |
Knowl. Manag. Aquat. Ecosyst.
Number 426, 2025
Riparian ecology and management
|
|
|---|---|---|
| Article Number | 14 | |
| Number of page(s) | 4 | |
| DOI | https://doi.org/10.1051/kmae/2025012 | |
| Published online | 21 May 2025 | |
Short Communication
The complete mitochondrial genome of the Mekong Giant Salmon Carp (Aaptosyax grypus): a tool to conserve an emblematic and elusive megafish
1
Centre de Recherche sur la Biodiversité et l’Environnement, CRBE UMR 5300 UPS-CNRS-IRD-INP, Université Paul Sabatier-Toulouse III, 118 routes de Narbonne, CEDEX 4, 31062 Toulouse, France
2
Department of Biology and Global Water Center, University of Nevada, 1664 N. Virginia Street, Reno, NV 89557, USA
3
Faculty of Agriculture, Svay Rieng University, National Road No.1, Sangkat Chek, Svay Rieng City, Cambodia
4
Inland Fisheries Research and Development Institute, Fisheries Administration, # 186, Preah Norodom Blvd., Sangkat Tonle Bassak, Khan Chamkar Mon, Phnom Penh 120101, Cambodia
5
Center of Excellence on Fisheries and Aquatic Sciences for Sustainability, Faculty of Fisheries and Aquaculture, Royal University of Agriculture, Sangkat Dangkor, Khan Dangkor, P.O. Box 2696, Phnom Penh 120501, Cambodia
* Corresponding author: celine.condachou@univ-tlse3.fr
Received:
7
February
2025
Accepted:
29
April
2025
The giant salmon carp is a Critically Endangered megafish from the Mekong River. Despite its emblematic species status, it is rarely observed, and less than 30 confirmed specimens have been recorded since the original species description in 1991. Here, we benefitted from the latest discovery to collect tissues for mitogenome sequencing. We provide the full mitogenome of Aaptosyax grypus, which is 16601 bp in length and codes for 13 genes. This mitogenome sequence will help resolve the phylogenetic placement of this emblematic species. It also provides the 12S gene reference sequence commonly used in environmental DNA (eDNA) studies and thus opens opportunities for the detection of this elusive species using eDNA, making it possible to investigate the spatial distribution of the species and develop strategies to conserve this Critically Endangered megafish.
Key words: Giant salmon carp / environmental DNA / phylogeny / conservation / enigmatic species / flagship species / critically endangered fish
© C. Condachou et al., Published by EDP Sciences 2025
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License CC-BY-ND (https://creativecommons.org/licenses/by-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. If you remix, transform, or build upon the material, you may not distribute the modified material.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License CC-BY-ND (https://creativecommons.org/licenses/by-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. If you remix, transform, or build upon the material, you may not distribute the modified material.
The giant salmon carp (Aaptosyax grypus) is among the most elusive freshwater fishes. Although large (up to 1.30 metre and >30 kg), this megafish was formally described only 35 years ago (Rainboth, 1991), and no more than 30 adult specimens have been recorded since its original description (Chan et al., 2024). It is part of an isolated clade from the Cyprinidae family, with no congeneric species or close relatives, and the lack of genetic data makes its phylogenetic placement uncertain within the Cyprinidae (Gaubert et al., 2009; Yang et al., 2015). This species is moreover considered Critically Endangered by the IUCN (Vidthayanon, 2011). It was suspected to be extinct because no specimens were recorded from 2005 to 2020, until the recent incidental captures by gillnets of three specimens in the Cambodian Mekong and one of its major tributaries (Chan et al., 2024). Therefore, the species still exists, providing an opportunity for protection (Chan et al., 2024), but its marked rarity hampers conservation actions, which as a foundation requires knowledge about its spatial distribution. The sequence presented here could help solve the phylogenetic placement of the species and will also enable the detection of the species through environmental DNA (eDNA) techniques, therefore allowing the assessment of the current distribution of the species to set baselines for future conservation action.
The total mitochondrial DNA was isolated from muscle tissue sampled on the adult specimen collected by local fishermen from the Sesan reservoir in Sesan District, Stung Treng Province, Cambodia, on June 08, 2022. The specimen was deposited at the Fish Collection Room of the Inland Fisheries Research & Development Institute (IFReDI), Fisheries Administration, Cambodia (voucher code CAMB-AGRY01). It was an adult, weighing 5 kg and 72.5 cm in total length, leaving no doubt about the species identification (Fig. 1). Additional DNA samples are available upon request.
Total genomic DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA, cat#69506) following manufacturer’s instructions. The concentration of the extracted DNA was 35.6 ng/μL. Genomic DNA library preparation was performed using a NEBNext Ultra™ II FS DNA Library Prep kit for Illumina (NEB #E7805S/L, #E6177S/L), optimised to create short fragment of approximately 300 pb. The libraries were sequenced on Illumina Novaseq 6000 using SP reagent kit v1.5 (500 cycles; cat#20028402) using the Texas A&M Agrilife Genomics and Bioinformatics Sequencing Core facility. Necessary quality control measures were performed at the sequencing centre prior to sequencing. Briefly, an Agilent Fragment Analyser was used to check fragment size and quality. Raw Illumina data were pre-processed with fastp to remove adapters and low-quality reads (Chen, 2023). Putative mitochondrial reads were skimmed and assembled de novo using GetOrganelle (Jin et al., 2020). The resulting assembly was annotated using MitoFish (Iwasaki et al., 2013). We then manually inspected all start and stop codon and curated all annotations in Geneious R9 (Kearse et al., 2012). The mitogenome was visualised using OGDRAW (Greiner et al., 2019).
The complete mitogenome of A. grypus is 16601 bp in length (GenBank accession no. PV069721), comprising 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a 945 bp long control region or D-loop (Fig. 2). The majority of genes were found on the H strand, except for ND6, tRNAGlu, tRNAPro, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, and tRNASer, which were encoded on the L strand. All protein-coding genes started with an ATG codon except for the cytochrome c oxidase subunit I (COXI) genes which started with the GTG codon. Seven TAA stop codon and one TAG stop codon were identified. Five incomplete stop codons were found (ND2, COXII, ND3, ND4, CytB). The GC% of the mitogenome of the studied species was found to be 43.1. D-loop was found highly AT rich with 67.3% AT content.
The complete mitochondrial genome produced in the present study represents the most complete DNA sequence for A. grypus. It might be a useful tool for the phylogenetic placement of the species in the Cyprinids phylogeny. Its current placement is uncertain, and A. grypus was proposed as a basal member of the Barbinae in the Cyprinids supertree generated by Gaubert et al. (2009) from multiple morphological and genetic sources. In contrast, Yang et al. (2015) consider A. grypus as part of the Cyprinini based on molecular data. Still, this last study indicates that the taxonomic position of A. grypus remains uncertain due to the paucity of available genetic data (only a single Cytb sequence was available). We therefore believe that integrating the full A. grypus mitogenome to future phylogenetic studies will ease the placement of this enigmatic species.
The full mitogenome also opens opportunities for the detection of A. grypus in the natural environment using environmental DNA techniques, which are now recognised as a fast and efficient technique for fish inventories (Takahashi et al., 2023). The 12S sequence provided here will allow the inclusion of A. grypus in the eDNA reference libraries using the most common and recognised primers for fishes, such as Teleo1 and Mifish (Polanco et al., 2021). It also provides the opportunity to develop species-specific markers designed for quantitative eDNA techniques such as qPCR or digital PCR (Jerde et al., 2021). Leveraging our ability to detect the presence and even quantify the abundance of A. grypus through eDNA techniques would be an asset for the conservation of this elusive species. It would allow us to map the species’ spatial distribution range and design targeted conservation in these areas. The complete mitogenome provided here, therefore, unlocks the development of conservation approaches to this emblematic but Critically Endangered species.
Acknowledgements
This study was funded by the Wonders of the Mekong project, supported by the United States Agency for International Development (USAID) (Cooperative Agreement #AID-OAA-A-16-00057). We are grateful to the Inland Fisheries Research and Development Institute of the Cambodian Fisheries Administration for their collaboration in exploring of the species. We express our gratitude to Mr Sokheng Chan, a former staff of the Inland Fisheries Research and Development Institute who passed away in 2024, for his efforts in exploring and tracking the presence of Aaptosyax grypus by building a strong network with the local fishermen.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of the NCBI database at https://www.ncbi.nlm.nih.gov under accession no. PV069721.
References
- Chan B, Hogan ZS, Grant MI, Chhuoy S, Chhut C, Heng K, Brosse S. 2024. Rediscovery and future approaches to conservation of the elusive giant salmon carp Aaptosyax grypus, a Critically Endangered megafish in the Mekong. Biol Conserv 299: 110814. [CrossRef] [Google Scholar]
- Chen S. 2023. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2: e107. [CrossRef] [Google Scholar]
- Gaubert P, Denys G, Oberdorff T. 2009. Genus‐level supertree of Cyprinidae (Actinopterygii: Cypriniformes), partitioned qualitative clade support and test of macro‐evolutionary scenarios. Biol Rev 84: 653–689. [CrossRef] [PubMed] [Google Scholar]
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res 47: W59–W64. [CrossRef] [PubMed] [Google Scholar]
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M. et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol 30: 2531–2540. [CrossRef] [PubMed] [Google Scholar]
- Jerde CL, Mahon AR, Campbell T, McElroy ME, Pin K, Childress JN, Armstrong MN, Zehnpfennig JR, Kelson SJ, Koning AA, Ngor PB, Vanna N, So N, Chandra S, Hogan ZS. 2021. Are genetic reference libraries sufficient for environmental DNA metabarcoding of mekong river basin fish? Water 13: 1767. [CrossRef] [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol 21: 1–31. [CrossRef] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [CrossRef] [PubMed] [Google Scholar]
- Polanco A, Richards E, Flück B, Valentini A, Altermatt F, Brosse S, Walser JC, Eme D, Marques V, Manel S, Albouy C, Dejean T, Pellissier L. 2021. Comparing the performance of 12S mitochondrial primers for fish environmental DNA across ecosystems. Environ DNA 3: 1113–1127. [CrossRef] [Google Scholar]
- Rainboth WJ. 1991. Aaptosyax grypus, a new genus and species of large piscivorous cyprinids from the middle Mekong River. Japanese J Ichthyol 38: 231–237. [Google Scholar]
- Takahashi M, Saccò M, Kestel JH, Nester G, Campbell MA, van der Heyde M, Heydenrych MJ, Juszkiewicz DJ, Nevill P, Dawkins KL, Bessey C, Fernandes K, Miller H, Power M, Mousavi-Derazmahalleh M, Newton JP, White NE, Richards ZT, Allentoft ME. 2023. Aquatic environmental DNA: a review of the macro-organismal biomonitoring revolution, Sci Total Environ 873: 162322. [Google Scholar]
- Vidthayanon C. 2011. Mekong Giant Salmon Carp Aaptosyax grypus [WWW Document]. IUCN Red List Threat Species 2011. URL http://dx.doi.org/10.2305/IUCN.UK2011-1.RLTS.T9A13090494.en [Google Scholar]
- Yang L, Sado T, Hirt MV, Pasco-Viel E, Arunachalam M, Li J, Wang X, Freyhof J, Saitoh K, Simons AM, Miya M, He S, Mayden RL. 2015. Phylogeny and polyploidy: resolving the classification of cyprinine fishes (Teleostei: Cypriniformes). Mol Phylogenet Evol 85: 97–116. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Condachou C, Hogan ZS, Chan B, Chhut C, Heng K, Chhuoy S, Brosse S. 2025. The complete mitochondrial genome of the Mekong Giant Salmon Carp (Aaptosyax grypus): a tool to conserve an emblematic and elusive megafish. Knowl. Manag. Aquat. Ecosyst., 426. 14. https://doi.org/10.1051/kmae/2025012
All Figures
 |
Fig. 1 The voucher specimen of Aaptosyax grypus used for full mitogenome sequencing. |
| In the text | |
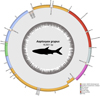 |
Fig. 2 Map of Aaptosyax grypus mitochondrial genome of 16601 bp representing 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a 945 bp long control region or D-loop. The annotated features of the outer circle are colored by their functional categories as shown in the legend (bottom right), and the inner circle indicates the GC content. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


