| Issue |
Knowl. Manag. Aquat. Ecosyst.
Number 421, 2020
|
|
|---|---|---|
| Article Number | 48 | |
| Number of page(s) | 8 | |
| DOI | https://doi.org/10.1051/kmae/2020040 | |
| Published online | 18 December 2020 | |
Research Paper
Potamopyrgus antipodarum (Gray, 1843) in Polish waters − its mitochondrial haplotype and role as intermediate host for trematodes
Potamopyrgus antipodarum (Gray, 1843) dans les eaux polonaises – son haplotype mitochondrial et son rôle d'hôte intermédiaire pour les trématodes
1
Faculty of Biological and Veterinary Sciences, Department of Invertebrate Zoology and Parasitology, Nicolaus Copernicus University in Torun, Lwowska 1, Torun 87-100, Poland
2
Institute of Environmental Sciences, Jagiellonian University, Gronostajowa 7, Krakow 30-387, Poland
3
Institute of Zoology and Biomedical Research, Jagiellonian University, Gronostajowa 9, Krakow 30-387, Poland
4
Faculty of Biological and Veterinary Sciences, Department of Ecology and Biogeography, Nicolaus Copernicus University in Torun, Lwowska 1, Torun 87-100, Poland
* Corresponding author: ezbikow@umk.pl
Received:
2
August
2020
Accepted:
30
November
2020
The New Zealand mud snail (Potamopyrgus antipodarum (Gray, 1843)) is on the list of one hundred worst invasive species. Researchers point out that genetic variation between populations of P. antipodarum manifested in differences in life-history traits. The main objective of our investigation was to gain pioneer knowledge about mitochondrial haplotypes of P. antipodarum in Polish waters on the background of these haplotypes recorded in the world and confirmation of the main role of P. antipodarum in the life cycle of digenean trematodes. We examined 1000 individuals of P. antipodarum from five water bodies in three different parts of Poland for the presence of larval stages of digenean trematodes. For several randomly selected individuals we carried out DNA sequencing of the 16S ribosomal RNA gene as marker of this non-indigenous mollusk. Only one 16S rRNA haplotype of P. antipodarum was recorded in Polish waters, defined in this study as haplotype 1 which turned out to be the most widespread in Europe. Potamopyrgus antipodarum is a source of trematode metacercariae belonging mainly to the family Echinostomatidae. As a result, we can demonstrate that it plays a role as the second intermediate host of digenean trematodes in European waters.
Résumé
L'escargot de vase de Nouvelle-Zélande (Potamopyrgus antipodarum (Gray, 1843)) figure sur la liste des cent pires espèces envahissantes. Les chercheurs soulignent que la variation génétique entre les populations de P. antipodarum se manifeste par des différences dans les traits du cycle de vie. L'objectif principal de notre étude était d'acquérir des connaissances préliminaires sur les haplotypes mitochondriaux de P. antipodarum dans les eaux polonaises sur le fond de ces haplotypes enregistrés dans le monde et de confirmer le rôle principal de P. antipodarum dans le cycle de vie des trématodes digéniens. Nous avons examiné 1 000 individus de P. antipodarum provenant de cinq plans d'eau dans trois régions différentes de Pologne pour détecter la présence de stades larvaires de trématodes digéniens. Pour plusieurs individus sélectionnés au hasard, nous avons effectué le séquençage de l'ADN du gène ARN ribosomique 16S comme marqueur de ce mollusque non indigène. Un seul haplotype d'ARNr 16S de P. antipodarum a été enregistré dans les eaux polonaises, défini dans cette étude comme l'haplotype 1 qui s'est avéré être le plus répandu en Europe. Potamopyrgus antipodarum est une source de métacercaires trématodes appartenant principalement à la famille des Echinostomatidae. Nous pouvons ainsi démontrer qu'il joue un rôle en tant que deuxième hôte intermédiaire des trématodes digéniens dans les eaux européennes.
Key words: Freshwater snail / New Zealand mud snail / 16S rRNA / metacercariae / biological invasion
Mots clés : Escargot d'eau douce / escargot de vase de Nouvelle-Zélande / ARNr 16S / métacercaires / invasion biologique
© A. Stanicka et al., Published by EDP Sciences 2020
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License CC-BY-ND (https://creativecommons.org/licenses/by-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. If you remix, transform, or build upon the material, you may not distribute the modified material.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License CC-BY-ND (https://creativecommons.org/licenses/by-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. If you remix, transform, or build upon the material, you may not distribute the modified material.
1 Introduction
The New Zealand mud snail Potamopyrgus antipodarum (Gray, 1853) (Gastropoda, Tateidae) has been present in Europe for about 160 yrs (Boycott, 1936), while in Poland, its first occurrence was recorded almost 100 yrs ago (Urbański, 1935). Currently, this is a species with a worldwide distribution (Collado, 2014) that causes numerous ecological and economic problems (Alexandre da Silva et al., 2019). Potamopyrgus antipodarum has also been presented among the one hundred worst invasive species in Europe (Nentwig et al., 2018). The latest models of potential distribution and areas susceptible to the invasion of P. antipodarum in South America and worldwide, regardless of positive and negative climate changes, show greater suitability of the environment, also outside the area of its current distribution (Alexandre da Silva et al., 2019).
One of the strategies that make the species an effective invader in a new area is parthenogenesis (i.e., asexual reproduction in which development of embryos occur without fertilization) (Jacobsen and Forbes, 1997). In the European populations, there is no evidence of sexual reproduction, although males of P. antipodarum have been found (Jacobsen and Forbes, 1997). The populations of parthenogenetic organisms can expand their range more efficiently while adapting to a new environment (Morgan-Richards et al., 2010). The parthenogenesis of P. antipodarum living outside native boundaries under favourable and stable conditions allows for a fast multiplication of specimens that become quantitative dominants among macroinvertebrates (Alonso and Castro-Díez, 2012). Städler et al. (2005) showed a marked divergence between the two European haplotypes of P. antipodarum, which means successful colonization by two distinct mitochondrial lineages. However, the samples of Polish origin have not been studied so far.
Ballast water is considered the main cause of the global spread of this invasive species (Alonso and Castro-Díez, 2008). However, the presence of the snail species in Polish water bodies as well as other non-endemic places, may result from other passive modes of its distribution in the spread of the individuals on birds' feathers or in fish intestines (Alonso and Castro-Díez, 2008). The success of P. antipodarum in inhabiting non-native areas is associated with a wide tolerance to different physicochemical conditions (e.g., pH, water temperature, dissolved oxygen), as well as the shell morphological adaptations and the lack of native enemies. For example, thanks to the solid operculum and strong shell, snails are able to survive in the digestive system of fish, as a result, these vertebrates are less effective as predators, and on the contrary, they can be the carrier of P. antipodarum (Alonso and Castro-Díez, 2012).
Very little has been proven about the use of P. antipodarum by parasites in its new areas (Larson and Krist, 2019). The snail is even considered as a potential biological control against some species of parasites such as bird schistosomes (Marszewska et al., 2018a). However, it should be taken into account that this snail species, reaching such huge densities (Dorgelo, 1987), may become a convenient host for local parasites. In the work of Kelly et al. (2009) a wide range of arguments is presented that nonindigenous animal species may be highly competent hosts for the indigenous ones.
The genetic structure of intermediate hosts of trematodes is of great interest to scientists (Hauswald et al., 2011; Tantrawatpan et al., 2020). The genetic variation between populations of P. antipodarum is reflected e.g. in differences in life-history traits (Jacobsen and Forbes, 1997). We examined specimens of this alien species from different localization in Poland to obtain pioneer knowledge about its mitochondrial haplotypes in the area of Central Europe, which we presented on the background of the recorded mitochondrial haplotypes in the world, and we checked the role of P. antipodarum as an intermediate host for digenean trematodes.
2 Material and methods
2.1 Research area and sampling
The samples were gathered in September 2018, that is, after months of the strongest release of cercariae from the first intermediate hosts (various species of freshwater snails) (Marszewska et al., 2018b; Cichy et al., 2019) for which P. antipodarum can play the role of the second intermediate host (Żbikowski and Żbikowska, 2009; Cichy et al., 2017). The study areas which we used for the research were water bodies from three different parts of Poland: western Poland − Lake Czaplino; central − Lake Iławskie and Lake Sosno; and southern Poland − a pond in Strzemieszyce Wielkie (a district of Dąbrowa Górnicza), as well as a bathing resort in the Valley of Three Ponds area in Katowice (Fig. 1). All of the lakes situated in the north part of Poland are natural water bodies, and the others are anthropogenic reservoirs that seem more attractive to P. antipodarum (Johnson et al., 2008). Also, the abiotic conditions of the environment are of great importance for the formation of mollusk populations (Larson et al., 2020), and therefore the values of physical and chemical parameters of water were measured during the study with the core sampler and a MultiLine P4 (WTW) Universal Pocket Sized Meter (Tab. 1). The sampling of P. antipodarum from a sandy bottom (up to a depth of 1.5 m) in each site was carried out using a metal sieve (mesh diameter 3 mm). Then the snails were transported to the laboratory in containers with lake water. The taxonomic affiliation of collected snails as P. antipodarum species was verified on the basis of morphological data (Piechocki and Wawrzyniak-Wydrowska, 2016).
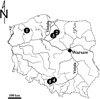 |
Fig. 1 Study sites in Poland, Central Europe: 1–Lake Czaplino (53°32′59″N, 16°14′59″E), 2–Lake Iławskie (53°35′37″N, 19°36′54″E), 3–Lake Sosno (53°20′15″N, 19°20′55″E), 4–pond in Strzemieszyce Wielkie (district of Dąbrowa Górnicza) (50°18′23″N, 19°19′24″E), 5–a bathing resort in the Valley of Three Ponds area in Katowice (50°14′32″N, 19°02′42″E). |
The mean values of conductivity and pH from the study sites.
2.2 Parasitological examination of snails
In each sample, we randomly isolated 200 individuals with a shell height of 4–6 mm from each research area for an autopsy. The snail shells were removed and the soft parts of the body were carefully examined for the presence of trematodes under a light microscope (Axio Lab.A1), whereas using the Axiocam 105 color camera and ZEN software, photographs and measurements were taken. The identification was made on the basis of the morphological characteristics of live parasite larvae. For this purpose, we used drawings, photos, and descriptions of parasites presented by various authors (Kanev, 1994; Gérard and Le Lannic, 2003; Faltýnková et al., 2007; Żbikowski and Żbikowska, 2009; Cichy and Żbikowska, 2016; Cichy et al., 2017).
The term “prevalence” is the percentage ratio of the number of infected individuals to the number of all collected P. antipodarum in a given water reservoir, calculated according to the formula: P [%] = n/N × 100 (P = prevalence, n = the number of infected individuals in a sample, N = the number of all collected individuals in a sample). The term “average intensity” is the average number of metacercariae recorded per infected individual, calculated according to the formula: AI = S/n (AI = average intensity, S = the sum of all larvae in a sample, n = the number of infected individuals in a sample).
2.3 DNA extraction, amplification and sequencing of P. antipodarum
DNA extraction was performed for five individuals of P. antipodarum from each research area, with the exception of Lake Sosno, in the case of which, 3 specimens were tested, using the Sherlock AX Kit (A & A Biotechnology, Poland). PCR generated a fragment of the 16S ribosomal RNA gene using the two primers, S1-Universal (5′-CGGCCGCCTGTTTATCAAAAACAT-3′) and S2-Potamo (5′-GTGGTCGAACAGACCAACCC-3′) (Städler et al., 2005). A PCR reaction of each sample was performed in a 20 µl reaction mixture, consisting of 3 µl of template DNA, 0.6 µl of each primer, 2 µl of 10× buffer, 13 µl of ddH2O, 0.6 µl of 20 mM dNTP (ThermoFisher Scientific, USA) and 0.2 µl of Taq-Polymerase (ThermoFisher Scientific, USA). PCR conditions consisted of 5 min initial denaturation at 92 °C, 30 s denaturation at 92 °C, followed by 60 s annealing at 55 °C, and 90 s elongation at 72 °C for 40 cycles followed by a final elongation step for 5 min at 72 °C. A 3 µl sample of PCR product was run on a 1.5% agarose gel for 30 min at 100 V to check DNA quality. PCR products were cleaned up by using Clean-up Kit (A&A Biotechnology, Poland). A sequencing reaction was performed in 10 µl of the reaction mixture, consisting of 2 µl of PCR product, 0.15 µl of primer, 1 µl of sequencing buffer (Brilliant Dye Terminator Sequencing Kit, Nimagen, The Netherlands), 5.85 µl of ddH2O and 1 µl of Terminator (Brilliant Dye Terminator Sequencing Kit, Nimagen, The Netherlands). The sequencing programme consisted of four steps: 1 min initial denaturation at 96 °C, followed by 10 s denaturation at 96 °C, 5 sec annealing at 55 °C, and 4 min elongation at 60 °C for 25 cycles. Sequencing products were cleaned up by using ExTerminator (A&A Biotechnology, Poland) and sequenced in one direction. The sequencing reactions were performed in the Genomed company (Warsaw, Poland). Sequences were deposited in GenBank with the following accession numbers: MK578223, MK578224, MK578225, MK578226, MK578227.
2.4 Alignment and statistical analyses
All available mitochondrial 16S ribosomal RNA (rRNA) sequences of P. antipodarum were downloaded from GenBank (Tab. 2). These sequences together with 23 newly obtained sequences were aligned by using MAFFT version 7 (Katoh et al., 2002; Katoh and Toh, 2008). A 16S dataset (436 bp length) comprises sequences from 16 countries (Tab. 2). To estimate the population genetic parameters (such as the number of haplotypes, polymorphic sites (S), nucleotide diversity (π), haplotype diversity (H)) calculations were performed in DnaSP v.5.10 (Librado and Rozas, 2009). Median Joining haplotype network (436 bp length of alignment) was performed in PopART (Bandelt et al., 1999). The haplotype networks were constructed from all haplotype sequences presented in each studied population. Haplotypes were shown as black circles, where the size of the circle represents the number of populations where they are present. Haplotypes are identified by numbers with # marks, white circles without a number indicate a hypothetical intermediate haplotype which is necessary to link observed haplotypes. Hatch marks in the network represent single mutations.
Haplotypes of Potamopyrgus antipodarum in certain localities based on 16S ribosomal RNA gene.
3 Results
3.1 Molecular examination
Based on all 16S rRNA sequences of P. antipodarum obtained from our samples, we observed that all of them belong to the same haplotype, defined in this study as haplotype 1 (Tab. 2, Fig. 2). Together with all available mitochondrial 16S ribosomal RNA sequences of P. antipodarum, we identified 22 distinct haplotypes. Haplotype 1 and 2 were most frequent, present in 11 and 6 countries, respectively. Apart from Poland, haplotype 1 was present in Germany, France, Hungary, Lithuania, Spain, Denmark, Italy, the Netherlands, Switzerland and the United Kingdom (UK), whereas haplotype 2 occurs in Estonia, the USA, Spain, Belgium, France and the UK (Tab. 2). Haplotypes from 3 to 17 and from 19 to 22 were restricted to water bodies of New Zealand, while haplotype 18 occurs in Tasmania (Australia). Population genetic parameters for P. antipodarum were S = 22; π = 0.00804; H = 1.000.
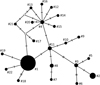 |
Fig. 2 Haplotype Median Joining network for the 16S ribosomal RNA haplotypes of Potamopyrgus antipodarum. Haplotypes are represented by black circles, the size of which is proportional to the number of populations in which a particular haplotype is present. Populations are listed in Table 2. Haplotypes are from #1 to #22; white circles without a number indicate a hypothetical intermediate haplotype linking observed haplotypes. Hatch marks in the network represent single mutations. |
3.2 Parasitological examination
We found three morphological types of metacercariae (Fig. SM 1, Tab. 3). They differed both in diameter and in the presence/absence of collar spines. The metacercaria of type I (from Lake Czaplino) was characterized by the outer diameter of cyst: 118 × 116 μm. Inside the cyst, we observed two suckers and we could not observe a stylet or collar spines. The metacercariae of type II were found in snails from Lake Iławskie and Pond in Strzemieszyce Wielkie, and they had the outer diameter of cysts: 149 (SE 0.9) × 148 (SE 0.7) μm and 150 (SE 0.5) × 148 (SE 0.5) μm, respectively. The metacercariae of type III recorded in Lake Sosno were much larger than the others (261 (SE 0.6) × 260 (SE 0.7) μm in diameter). All the larvae also had two rows of collar spines, but 4 spines at both edges of the collars were much larger than the other spines. The presence of these larvae was recorded in specimens from four out of the five investigated water bodies (Tab. 3). The prevalence ranged from 0.5% to 6%, and the average intensity from 1 to 2.5 larvae per one host snail (Tab. 3).
Infection of metacercariae in Potamopyrgus antipodarum.
4 Discussion
A single parthenogenetic individual of P. antipodarum can colonize a new area by itself (Schreiber et al., 1998). These populations consist of “snail clones” because only new introductions and/or mutations can increase the diversity of the gene pool (Weetman et al., 2006). As a result, snail populations living outside their native range are characterized by low genetic diversity (Alonso and Castro-Díez, 2012). All P. antipodarum used in our studies represented the same haplotype, despite the fact that they came from five water bodies which are located at a substantial distance from each other. Our data is the first demonstration of the presence of 16S rRNA haplotype in Poland. In the case of the whole Europe, there are several clonal lineages presumably created as a consequence of the long invasion history (Alonso and Castro-Díez, 2012). Our analysis showed that the haplotype found in our research is the most widespread in Europe.
Potamopyrgus antipodarum individuals from our study were infected only with metacercariae. In New Zealand, P. antipodarum is widely used by many species of digenean trematodes not only as the second but also as a strongly specific first intermediate host (Hechinger, 2012). For example, Hechinger (2012) listed 20 species of digenean trematodes for which P. antipodarum plays the role of an intermediate host in its native area, with the prevalence up to 7%. By contrast, outside the native range, pre-patent or patent infection (infections with sporocysts, rediae or immature cercariae) is extremely rare (Evans et al., 1981; Gérard and Le Lannic, 2003; Żbikowski and Żbikowska, 2009). Recorded cases of such digenean infections found in P. antipodarum are the result of an introduction from their native areas (Gérard et al., 2017). We recorded a low prevalence of snails infected with metacercariae, similar to our previous study presented by Cichy et al. (2017). Because too few morphological features have been observed to identify the species of metacercariae found in Lake Czaplino, we were unable to determine whether it posed a medical or veterinary threat. Other metacercariae detected in P. antipodarum from our study were identified to belong to the family Echinostomatidae (Kanev, 1994; Faltýnková et al., 2007; Cichy and Żbikowska, 2016). Given the characteristics of metacercariae, we suspect that these larvae represent the genus Echinoparyphium and/or Echinostoma (Kanev, 1994; Faltýnková et al., 2007; Cichy and Żbikowska, 2016). There are several species of Echinostoma (e.g. E. revolutum, E. paraulum) and Echinoparyphium (e.g. E. recurvatum, E. mordwilkoi) in Europe, which at the stage of metacercaria cannot be distinguished without molecular methods (Georgieva et al., 2014). Members of these genera are quite common in the environment and use common snail species as first or second intermediate hosts (Cichy and Żbikowska, 2016). Echinostomes are widespread flukes causing intestinal diseases and they are mainly important in wildlife diversity (Saijuntha et al., 2010).
In conclusion, our research indicates the presence of only one 16S rRNA haplotype of P. antipodarum in Poland. Potamopyrgus antipodarum outside its natural occurrence is used as the second intermediate host of digenean trematodes; however, the detection of the association between the mtDNA haplotype of P. antipodarum and metacercarial infection should be investigated in detail in the future.
Supplementary Material
Fig. SM 1. Metacercariae recorded in Potamopyrgusantipodarum from - A) Lake Czaplino, B) Lake Sosno, C) Pond in StrzemieszyceWielkie, Poland. Access here
Author contribution statement
Anna Stanicka and Elżbieta Żbikowska designed the study. Anna Stanicka, Kamila Stefania Zając, Dorota Lachowska-Cierlik, Anna Cichy and Janusz Żbikowski executed the study. Anna Stanicka and Kamila Stefania Zając analyzed and interpreted the data and wrote the manuscript. Elżbieta Żbikowska provided substantive contributions and critical review.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
We would like to thank Anna Cieplok, PhD of the University of Silesia in Katowice for help in collecting research material.
This project was supported by the grant of the National Science Centre, Poland No. 2017/25/N/NZ8/01345, and statutory funds from the Ministry of Science and Higher Education (PL) for the research programs of the Department of Invertebrate Zoology, Nicolaus Copernicus University in Torun, PL, including Grant No. 1200-B; also this work was supported by the Jagiellonian University, PL, including Grant No. K/ZDS/006320.
References
- Alexandre da Silva MV, Nunes Souza JV, Souza JRB, Vieira LM. 2019. Modelling species distributions to predict areas at risk of invasion by the exotic aquatic New Zealand mud snail Potamopyrgus antipodarum (Gray 1843). Freshw Biol 64: 1504–1518. [Google Scholar]
- Alonso A, Castro-Díez P. 2008. What explains the invading success of the aquatic mud snail Potamopyrgus antipodarum (Hydrobiidae, Mollusca)? Hydrobiologia 614: 107–116. [Google Scholar]
- Alonso Á, Castro-Díez P. 2012. The exotic aquatic mud snail Potamopyrgus antipodarum (Hydrobiidae, Mollusca): state of the art of a worldwide invasion. Aquat Sci 74: 375–383. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48. [CrossRef] [PubMed] [Google Scholar]
- Boycott AE. 1936. The habitats of fresh-water Mollusca in Britain. J Anim Ecol 5: 116–186. [Google Scholar]
- Cichy A, Żbikowska E. 2016. Atlas of Digenea developmental stages: The morphological characteristics and spread within the populations of freshwater snails from the Brodnickie Lakeland, Poland, NCU Press, Torun. [Google Scholar]
- Cichy A, Marszewska A, Parzonko J, Żbikowski J, Żbikowski E. 2017. Infection of Potamopyrgus antipodarum (Gray, 1843) (Gastropoda: Tateidae) by trematodes in Poland, including the first record of aspidogastrid acquisition. J Invertebr Pathol 150: 32–34. [CrossRef] [PubMed] [Google Scholar]
- Cichy A, Marszewska A, Ciapka P, Żbikowska E. 2019. Different thermal conditions of lakes affect host–parasite systems: a case study of Viviparus contectus (Millet, 1813) and digenean trematodes. Freshw Biol 42: 417–425. [Google Scholar]
- Clusa L, Ardura A, Gower F, Miralles L, Tsartsianidou V, Zaiko A, Garcia-Vazquez E. 2016. An easy phylogenetically informative method to trace the globally invasive Potamopyrgus mud snail from river's eDNA. PLoS One 11: e0162899. [Google Scholar]
- Collado GA. 2014. Out of New Zealand: molecular identification of the highly invasive freshwater mollusk Potamopyrgus antipodarum (Gray, 1843) in South America. Zool Stud 53: 1821. [Google Scholar]
- Dorgelo J. 1987. Density fluctuations in populations (1982–1986) and biological observations of Potamopyrgus jenkinsi in two trophically differing lakes. Hydrobiol Bull 21: 95–110. [CrossRef] [Google Scholar]
- Evans NA, Whitfield PJ, Dobson AP. 1981. Parasite utilization of a host community: the distribution and occurrence of metacercarial cysts of Echinoparyphium recurvatum (Digenea: Echinostomatidae) in seven species of mollusc at Harting Pond, Sussex. Parasitology 83: 1–12. [Google Scholar]
- Faltýnková A, Nasincová V, Kablásková L. 2007. Larval trematodes (Digenea) of the great pond snail, Lymnaea stagnalis (L.), (Gastropoda, Pulmonata) in Central Europe: a survey of species and key to their identification. Parasite 14: 39–51. [CrossRef] [EDP Sciences] [PubMed] [Google Scholar]
- Georgieva S, Faltýnková A, Brown R, Blasco-Costa I, Soldánová M, Sitko J, Scholz T, Kostadinova A. 2014. Echinostoma ‘revolutum’ (Digenea: Echinostomatidae) species complex revisited: species delimitation based on novel molecular and morphological data gathered in Europe. Parasit Vectors 7: 520. [Google Scholar]
- Gérard C, Le Lannic J. 2003. Establishment of a new host–parasite association between the introduced invasive species Potamopyrgus antipodarum (Smith) (Gastropoda) and Sanguinicola sp. Plehn (Trematoda) in Europe. J Zoology 261: 213–216. [CrossRef] [Google Scholar]
- Gérard C, Miura O, Lorda J, Cribb TH, Nolan MJ, Hechinger RF. 2017. A native-range source for a persistent trematode parasite of the exotic New Zealand mud snail (Potamopyrgus antipodarum) in France. Hydrobiologia 785: 115–126. [Google Scholar]
- Haase M. 2005. Rapid and convergent evolution of parental care in hydrobiid gastropods from New Zealand. J Evol Biol 18: 1076–1086. [Google Scholar]
- Hechinger RF. 2012. Faunal survey and identification key for the trematodes (Platyhelminthes: Digenea) infecting Potamopyrgus antipodarum (Gastropoda: Hydrobiidae) as first intermediate host. Zootaxa 3418: 1–27. [Google Scholar]
- Hauswald AK, Remais JV, Xiao N, Davis GM, Lu D, Bale MJ, Wilke T. 2011. Stirred, not shaken: genetic structure of the intermediate snail host Oncomelania hupensis robertsoni in an historically endemic schistosomiasis area. Parasit Vectors 4: 206. [Google Scholar]
- Jacobsen R, Forbes VE. 1997. Clonal variation in life-history traits and feeding rates in the gastropod, Potamopyrgus antipodarum: performance across a salinity gradient. Funct Ecol 11: 260–267. [Google Scholar]
- Johnson PTJ, Olden JD, Vander Zanden MJ. 2008. Dam invaders: impoundments facilitate biological invasions into freshwaters. Front Ecol Environ 8: 357–363. [Google Scholar]
- Kanev I. 1994. Life-cycle, delimitation and redescription of Echinostoma revolutum (Froelich, 1802) (Trematoda: Echinostomatidae). Syst Parasitol 125–144. [Google Scholar]
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9: 286–298. [CrossRef] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. [CrossRef] [PubMed] [Google Scholar]
- Kelly DW, Paterson RA, Townsend CR, Poulin R, Tompkins DM. 2009. Parasite spillback: a neglected concept in invasion ecology? Ecology 90: 2047–2056. [Google Scholar]
- Klymus KE, Marshall NT, Stepien CA. 2017. Environmental DNA (eDNA) metabarcoding assays to detect invasive invertebrate species in the Great Lakes. PLOS One 12: e0177643. [Google Scholar]
- Larson MD, Krist AC. 2019. Trematode prevalence and an invasive freshwater snail: fewer infections and parasites likely contribute to the success of an invasive snail. Biol Invasions 48: 275. [Google Scholar]
- Larson MD, Dewey JC, Krist AC. 2020. Invasive Potamopyrgus antipodarum (New Zealand mud snails) and native snails differ in sensitivity to specific electrical conductivity and cations. Aquat Ecol 54: 103–117. [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [CrossRef] [PubMed] [Google Scholar]
- Marszewska A, Cichy A, Bulantová J, Horák P, Żbikowska E. 2018a. Potamopyrgus antipodarum as a potential defender against swimmer's itch in European recreational water bodies-experimental study. PeerJ 6: e5045. [CrossRef] [PubMed] [Google Scholar]
- Marszewska A, Strzała T, Cichy A, Dąbrowska GB, Żbikowska E. 2018b. Agents of swimmer's itch-dangerous minority in the Digenea invasion of Lymnaeidae in water bodies and the first report of Trichobilharzia regenti in Poland. Parasitol Res 117: 3695–3704. [Google Scholar]
- Morgan-Richards M, Trewick SA, Stringer IAN. 2010. Geographic parthenogenesis and the common tea-tree stick insect of New Zealand. Mol Ecol 19, 1227–1238. [CrossRef] [PubMed] [Google Scholar]
- Nentwig W, Bacher S, Kumschick S, Pyšek P, Vilà M. 2018. More than “100 worst” alien species in Europe. Biol Invasions 20: 1611–1621. [Google Scholar]
- Piechocki A, Wawrzyniak-Wydrowska B. 2016. Guide to freshwater and marine Mollusca of Poland. Poznan: Bogucki, 280 p. [Google Scholar]
- Saijuntha W, Sithithaworn P, Andrews RH. 2010. Genetic differentiation of Echinostoma revolutum and Hypodereaum conoideum from domestic ducks in Thailand by multilocus enzyme electrophoresis. J Helminthol 84: 143–148. [CrossRef] [PubMed] [Google Scholar]
- Schreiber ESG, Glaister A, Quinn GP, Lake PS. 1998. Life history and population dynamics of the exotic snail Potamopyrgus antipodarum (Prosobranchia: Hydrobiidae) in Lake Purrumbete, Victoria, Australia. Mar Freshwater Res 49: 73. [CrossRef] [Google Scholar]
- Städler T, Frye M, Neiman M, Lively CM. 2005. Mitochondrial haplotypes and the New Zealand origin of clonal European Potamopyrgus, an invasive aquatic snail. Mol Ecol 14: 2465–2473. [CrossRef] [PubMed] [Google Scholar]
- Tantrawatpan C, Saijuntha W, Bunchom N, Suksavate W, Pilap W, Walalite T, Agatsuma T, Tawong W, Sithithaworn P, Andrews RH, Petney TN. 2020. Genetic structure and geographical variation of Bithynia siamensis goniomphalos sensu lato (Gastropoda: Bithyniidae), the snail intermediate host of Opisthorchis viverrini sensu lato (Digenea: Opisthorchiidae) in the Lower Mekong Basin revealed by mitochondrial DNA sequences. Int J Parasitol 50: 55–62. [Google Scholar]
- Urbański J. 1935. Dwa ciekawe gatunki ślimaków w Wielkopolsce. Wydawnictwa Okręgowej Komisji Ochrony Przyrody w Poznaniu 5: 108–114. [Google Scholar]
- Weetman D, Hauser L, Carvalho GR. 2006. Heterogeneous evolution of microsatellites revealed by reconstruction of recent mutation history in an invasive apomictic snail, Potamopyrgus antipodarum . Genetica 127: 285–293. [CrossRef] [PubMed] [Google Scholar]
- Żbikowski J, Żbikowska E. 2009. Invaders of an invader-trematodes in Potamopyrgus antipodarum in Poland. J Invertebr Pathol 101: 67–70. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Stanicka A, Zając KS, Lachowska-Cierlik D, Cichy A, Żbikowski J, Żbikowska E. 2020. Potamopyrgus antipodarum (Gray, 1843) in Polish waters − its mitochondrial haplotype and role as intermediate host for trematodes. Knowl. Manag. Aquat. Ecosyst., 421, 48.
All Tables
Haplotypes of Potamopyrgus antipodarum in certain localities based on 16S ribosomal RNA gene.
All Figures
 |
Fig. 1 Study sites in Poland, Central Europe: 1–Lake Czaplino (53°32′59″N, 16°14′59″E), 2–Lake Iławskie (53°35′37″N, 19°36′54″E), 3–Lake Sosno (53°20′15″N, 19°20′55″E), 4–pond in Strzemieszyce Wielkie (district of Dąbrowa Górnicza) (50°18′23″N, 19°19′24″E), 5–a bathing resort in the Valley of Three Ponds area in Katowice (50°14′32″N, 19°02′42″E). |
| In the text | |
 |
Fig. 2 Haplotype Median Joining network for the 16S ribosomal RNA haplotypes of Potamopyrgus antipodarum. Haplotypes are represented by black circles, the size of which is proportional to the number of populations in which a particular haplotype is present. Populations are listed in Table 2. Haplotypes are from #1 to #22; white circles without a number indicate a hypothetical intermediate haplotype linking observed haplotypes. Hatch marks in the network represent single mutations. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


