Fig. 1
Download original image
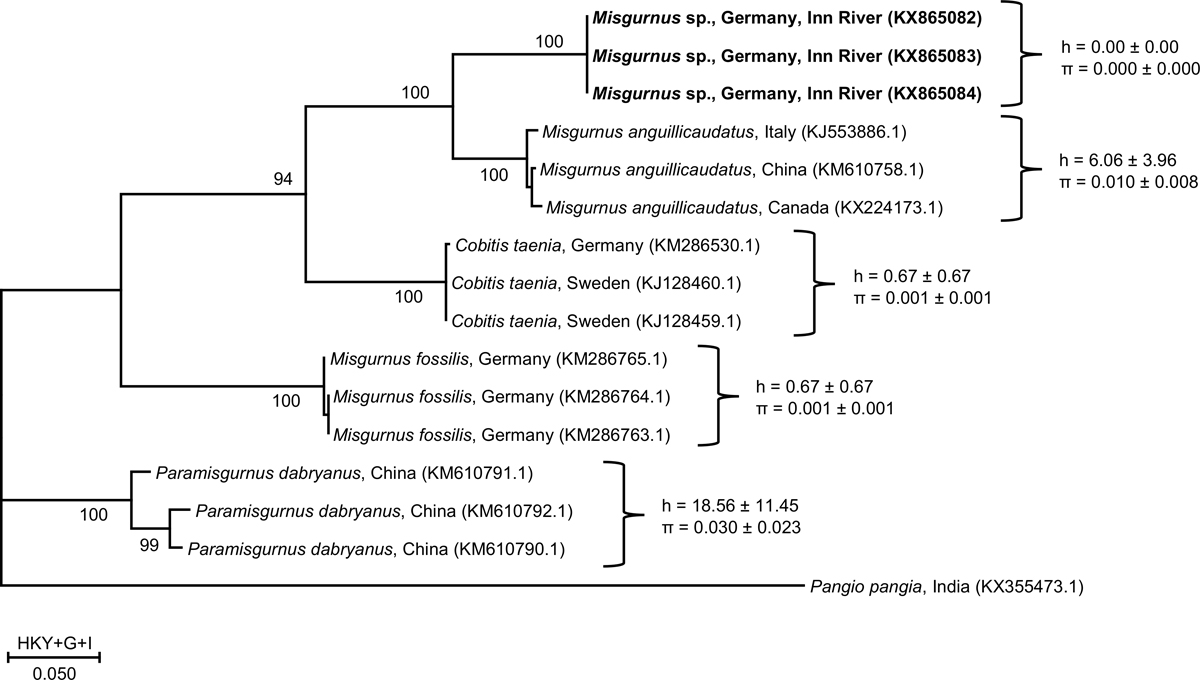
Maximum likelihood (ML) estimates of phylogenetic relationships of Misgurnus sp., P. dabryanus and Cobitis taenia COI sequences (622 bp). Nodes are labeled with the highest bootstrap support by ML (1000 replications). In bold: Sequences of the identified and sequenced specimen from the oxbow at River Inn, southern Germany. All branches had bootstrap support above 60%. The scale bar shows the best fit model chosen based on the Bayesian information criterion approach (Hasegawa–Kishino–Yano model with discrete Gamma distribution, HKY+G+I, above), and substitutions/site (below). In brackets, the current GenBank accession numbers of the published sequences used for this analysis. Values beside curly brackets indicate nucleotide (π) haplotype (h) and diversity.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


