| Issue |
Knowl. Manag. Aquat. Ecosyst.
Number 421, 2020
Topical issue on Crayfish
|
|
|---|---|---|
| Article Number | 50 | |
| Number of page(s) | 10 | |
| DOI | https://doi.org/10.1051/kmae/2020041 | |
| Published online | 23 December 2020 | |
Research Paper
Invasion and distribution of the redclaw crayfish, Cherax quadricarinatus, in Martinique
1
DEAL Direction de l’Environnement, de l’Aménagement et du Logement Route de la Pointe de Jaham - BP7212, Schœlcher 97274, Martinique
2
ODE Office De l'Eau, 7 Avenue Condorcet, Fort-de-France, Martinique
3
Laboratoire Ecologie et Biologie des Interactions, UMR CNRS 7267 Equipe Ecologie Evolution Symbiose, 5 rue Albert Turpin, Poitiers Cedex, France
4
Groupe BIOSPHERES, Université des Antilles, Campus de Schoelcher, Schoelcher 97200, Martinique
5
Centre for Integrative Ecology, School of Life and Environmental Sciences, Deakin University, Geelong, Victoria 3220, Australia
6
Department of Biological and Environmental Sciences, University of Gothenburg, PO Box 463, SE-405 31, Sweden
* Corresponding author: thmsbaudry@gmail.com
Received:
2
October
2020
Accepted:
30
November
2020
The redclaw crayfish, Cherax quadricarinatus, was introduced to Martinique Island for aquaculture purposes in 2004, in an attempt to revitalize the freshwater crustacean aquaculture sector. In 2015, three wild populations were discovered during an electrofishing survey on fish diversity. In 2018, a specific crayfish survey was performed at night using spotlighting and baited traps at 34 sites throughout the island. The species was mostly found in the center and northern part of the island, specifically, a total of 105 specimens were captured in eight streams and five closed water bodies. We sequenced a 491 base-pair fragment of the COI gene to understand the invasion history and pathway from the presumed source population at the Mangatal hatchery. Among the eight haplotypes found, three were dominant, of which, two occurred in the Mangatal hatchery. As crayfish are sold alive, there is a high risk of further human-mediated introductions across the island hydrographic basins. Thus, the distribution of this species could rapidly expand throughout Martinique freshwater ecosystems, with ecological impacts on native communities yet to be determined and requiring urgent investigation.
Résumé
L'écrevisse à pinces rouges, Cherax quadricarinatus, a été introduite en Martinique à des fins aquacoles en 2004, dans le but de revitaliser le secteur de l'aquaculture des crustacés d'eau douce. En 2015, trois populations sauvages ont été découvertes lors d'une étude sur la diversité des poissons par pêche électrique. En 2018, une étude spécifique sur les écrevisses a été réalisée de nuit à l'aide de lampes torche et de pièges appâtés sur 34 sites de l'île. L'espèce a été trouvée principalement dans le centre et le nord de l'île, plus précisément, un total de 105 spécimens a été capturés dans huit cours d'eau et cinq plans d'eau. Nous avons séquencé un fragment de 491 paires de bases du gène COI afin de comprendre l'histoire et le cheminement de l'invasion à partir de la population source présumée de l'écloserie Mangatal. Parmi les huit haplotypes trouvés, trois étaient dominants, dont deux dans l'écloserie de Mangatal. Comme les écrevisses sont vendues vivantes, il existe un risque élevé de nouvelles introductions d'origine humaine dans les bassins hydrographiques de l'île. Ainsi, la distribution de cette espèce pourrait rapidement s'étendre à l'ensemble des écosystèmes d'eau douce de la Martinique, avec des impacts écologiques sur les communautés indigènes qui restent à déterminer et qui nécessitent une investigation urgente.
Key words: Biodiversity hotspot / COI / freshwater / invasive species / parastacidae / wild detection
Mots clés : Hotspot de biodiversité / COI / eau douce / espèces envahissantes / parastacidae / détection sauvage
© T. Baudry et al., Published by EDP Sciences 2020
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License CC-BY-ND (https://creativecommons.org/licenses/by-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. If you remix, transform, or build upon the material, you may not distribute the modified material.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License CC-BY-ND (https://creativecommons.org/licenses/by-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. If you remix, transform, or build upon the material, you may not distribute the modified material.
1 Introduction
The introduction of non-indigenous species outside of their native environmental ranges constitutes a major threat to biological diversity, with significant negative impacts on their native counterparts and their ecosystems (Seebens et al., 2017). Exotic or alien species are found worldwide and among all the branches of the tree of life (Seebens et al., 2017; Blackburn et al., 2019). Among these species, the ones with the highest economic value are widely represented (Mcquaid and Arenas, 2009). The redclaw crayfish, also known as the tropical blue crayfish, Cherax quadricarinatus (Von Martens, 1868), is a striking example. Native to northern Australia and southern New Guinea (Austin, 1996; Bláha et al., 2016), C. quadricarinatus is popular for aquaculture and the aquarium trade because it readily breeds in captivity and grows fast in tropical climates (Jones, 1990; Patoka et al., 2014; Azofeifa-Solano et al., 2017). It has been deliberately translocated for aquaculture purposes or as an ornamental species to a number of countries, many with an equatorial or Mediterranean climate, including: Costa Rica (Azofeifa-Solano et al., 2017), Czech Republic (Patoka et al., 2015), Ecuador (Romero and Jimenez, 2002), Hungary (Weiperth et al., 2018), Indonesia (Patoka et al., 2018), Israel (Karplus et al., 1998; Snovsky and Galil, 2011), Kazakhstan (Uderbayev et al., 2017), Malaysia (Naqiuddin et al., 2016), Mexico (Bortolini et al., 2007), Russia (Vodovsky et al., 2017), Singapore (Ahyong and Yeo, 2007), Slovenia (Jaklič and Vrezec, 2011), South Africa (de Moor, 2002; Nunes et al., 2017), Taiwan (Hsieh et al., 2016), Ukraine (Kotovska et al., 2016), the United States of America (Morningstar et al., 2020), Zambia (Lodge et al., 2012) and Zimbabwe (Marufu et al., 2018). Similar to the Louisiana red swamp crayfish, Procambarus clarkii (Oficialdegui et al., 2020), live importation of redclaw crayfish has allowed the species to colonize new and often natural aquatic environments either through accidental escapes from hatcheries or ponds, or through direct human-mediated introductions. Because of its broad tolerance regarding water quality, a high reproductive rate and dispersal capacity it has a high invasion potential (Karplus et al., 1998; de Moor, 2002; Tropea et al., 2010), allowing the species to rapidly colonize entire river catchments.
Cherax quadricarinatus was first introduced to Martinique into aquaculture facilities in 2004 from Cuba, according to anecdotal information, in order to reinvigorate the aquaculture industry in the island, which was losing momentum as a result of ecotoxicological problems (in particular Macrobrachium rosenbergii). Also, according to anecdotal information, individuals were brought in one time and shared between farmers, but most of them have ceased crayfish farming because pesticide rate in their ponds exceeds limit for the sale of edible products. Because of its highly developed hydrographic networks and its tropical climate, Martinique possesses an optimal environment for this species. Since its introduction, and after escapes or intentional releases, this crayfish is now well established in three streams and one reservoir, according to observations from recent electrofishing campaigns conducted by government agencies, the Direction de l'Environnement de l'Aménagement et du Logement (DEAL Martinique) and the Office de l'Eau (ODE Martinique), within the framework of the EU Water Framework Directive, aiming at enhancing our knowledge on freshwater biodiversity (2013/2015/2016) (DEAL Martinique and ODE Martinique, 2016). Even if fishing is prohibited by law in Martinican freshwater rivers (R02-2017-12-28-003), it is carried out illegally, but the pressure on C. quadricarinatus, although in high demand by local fishermen, is far too low to limit population growth and dispersal. According to fishermen, populations are now found in high densities and the presence of this invasive species may therefore be impacting on the environment and native freshwater communities.
The objectives of this study were: (1) to confirm the taxonomic status of the specimens caught during the surveys by Cytochrome c Oxydase subunit I (COI) barcoding, (2) to update the distribution map of C. quadricarinatus in Martinique through trapping and nocturnal surveys in rivers of the island and (3) to understand its invasion pathways through the Martinican territory.
2 Material and methods
2.1 Study area
Martinique is a French volcanic island of 1128 km2 belonging to the Lesser Antilles, in the eastern Caribbean Sea (Fig. 1). The region is subject to a tropical climate with two main seasons: the “Carême” and the “Hivernage” and two inter-seasons. The “Carême” corresponds to the dry season, from February to April, with sunny and not very rainy weather with maximum temperatures around 30 °C. The “Hivernage”, lasting from July to November, is the rainy season, with the occurrence of storms or hurricanes. Temperature can reach up to 32 °C and annual precipitation range from 970 mm in the south, more gentle part of the island, to 6000 mm in the mountainous, more tropical northern part of the island (DEAL Martinique et al., 2018).
The Martinican hydrographic network is well developed due to the high rainfall and includes 70 catalogued major streams, fed by numerous small tributaries, both permanent and semi-permanent (Fig. 1) (DEAL Martinique et al., 2018). This provides Martinique with rich terrestrial and inland aquatic ecosystems, with a great diversity of fauna and flora, giving it a high rate of endemism within the West Indies (DEAL Martinique et al., 2018).
 |
Fig. 1 Cartography of the sampling sites of Cherax quadricarinatus together with the presence sites of this species in Martinique Island during 2018 field campaign. Sampling was carried out on permanent streams and in closed water bodies. Cartography was prepared using QGIS 2.18 (Las Palmas) software (QGIS Development Team 2016). The shape of Martinique Island was imported from the database IGN and the streams were found in BD Carthage and BD Topo. |
2.2 Field surveys
A total of 34 sites were surveyed between the 2nd of March and the 20th of May 2018 (during the Carême and the inter-season), including 20 rivers or streams and five closed water bodies (non-functional hatchery, ornamental ponds or water retention pond) throughout Martinique (Fig. 1). Four sites were sampled in the Lezarde River, the largest river in Martinique. The sampled sites were targeted according to the morphology of the stream (mean depth >20 cm for traps) and on the advice from local fishermen. We also included the four sites where C. quadricarinatus had been previously detected: Oman, Manzo, Petit Galion, Lezarde Site 2 (Tab. 1).
Sampling was done using four large baited-traps (80 cm length × 25 cm width × 25 cm height, with mesh of 1 cm × 1 cm and a single cone-shaped inlet) baited with cassava. Each site was search by spotlight after nightfall, for 20 min, on a transect of 100 m along the bank. After that, the traps were placed on stream beds or water holes (around 10 m from each other) and collected the next morning. Two trapping sessions were conducted at each site on two successive days. The site was considered as positive if at least one crayfish was caught during one of the two capture sessions or if crayfish were observed using spotlighting surveys at night.
Upon capture, individual crayfish were sexed, and total length was measured from the rostrum to the telson (to the closest 0.1 cm). One walking leg was collected from each captured individual and subsequently stored in 90% ethanol, in order to perform further analysis (DNA extraction, PCR and COI sequencing) to confirm the taxonomic status of crayfish in Martinique. The collected individuals were euthanised and not returned to the streams according to French law on invasive species (law n° 2016–1087 of August 8, 2016).
Summary of the 34 sites surveyed during the study, including 20 rivers (29 sites) and 5 closed water bodies (non-functional hatchery, ornamental ponds or water retention pond).
2.3 Samples collection and DNA extraction
A total of 105 individuals were analysed: 84 caught in the wild and 21 individuals were bought at the Mangatal Hatchery (Tab. 2). DNA extraction was performed from a fragment of leg muscle tissue using the Qiagen DNeasy® Blood & Tissue Kit protocol. The tissue was first dried, put in 180 μL ATL buffer and crushed with a sterile pestle. Twenty μL proteinase K was added to each tube and samples were then vortexed for 15 s before incubation at 56 °C until total lysis of tissue. Each sample was filled with 200 μL AL buffer and 200 μL of 100% EtOH. Samples were vortexed thoroughly, pipetted into a DNeasy Mini spin column and centrifuged at 6000 g for 1 min. Two washing steps were done on the column with AW1 and AW2 buffers. Finally, DNA was eluted by adding 200 μL Buffer AE and stored at −20 °C until further analysis. The DNA concentration was measured by Nanodrop® ND 100 spectrometer (Nanodrop Technologies, Wilmington, USA).
Inventory of the 111 red claw crayfish COI sequences used to carry out the haplotype network analysis.
2.4 COI mtDNA analysis
A fragment of COI gene was amplified by PCR using the “universal” primers for invertebrates (LCO1490 and HCO2198) previously designed by Folmer et al. (1994). For each individual, PCR was performed following the protocol in Chucholl et al. (2015): 2.5 min at 95 °C for the initial denaturing step, followed by 35 cycles of 45 s at 95 °C; 1 min at 48 °C and 1 min at 72 °C. The final elongation step was 10 min at 72 °C. Amplified products were purified and 1/10 diluted before sequencing on an ABI PRISM 3130xl automated sequencer (PE Applied BioSystems, Foster City, USA). Both forward and reverse primers were used for sequencing, and the resulting sequenced fragments were aligned and edited using Sequencher® (version 5.4, 2016) formatted with the reference insertion of Mendeley. The taxonomic assignment was assessed with BLASTn 2.10.1 (Madden, 2002) comparing COI sequences to the non-redundant database of NCBI (Version August 2020). A multiple alignment was performed with ClustalW implemented in BioEdit version 7.2.5 (Hall, 1999). This was then trimmed with GBLOCKS version 0.91b (Talavera and Castresana, 2007) to remove ambiguously aligned regions, and to maximize the resulting alignment length, we used the less stringent options to obtain a 491 nucleotides alignment length. All the newly generated sequences were deposited on NCBI GenBank under the accession numbers MW300737 to MW300837. We also added available sequences from GenBank comprising one sequence from Australia (DQ006294.1), one from Malaysia (NC_022937.1) and four from Indonesia (KX377345.1 to KX377348.1). A haplotype network was constructed based on the polymorphic sites of the trimmed COI sequences using the Median-Joining method implemented in PopART software version 1.7 (Leigh and Bryant, 2015) and genetic distance among haplotype and nucleotide diversity were also calculated with the same software.
3 Results
3.1 Taxonomic status
All crayfish sampled in Martinique belonged to the species C. quadricarinatus with a percentage of identity of COI sequences of 98.98 ± 0.34 with C. quadricarinatus sampled in Malaysia (Accession number: KF649850.1) (Tan et al., 2015), 99.19% ± 0.34 with those from Indonesia (Accession number: KX377345.1) (Patoka et al., 2016) and 99.63% ± 0.35 with those from Australia (Accession number: DQ006294.1) (Shull et al., 2005).
3.2 Distribution of cherax quadricarinatus
Among the 34 sites surveyed, crayfish were detected at 16 sites including eight streams and five closed water bodies, with C. quadricarinatus being recorded at ten new sites (six streams: Lorrain, Coulisses, Petite Riviere Site 1, Grand Galion, Petite Lezarde and Cacaos, and four closed water bodies: Chateau Gaillard, Jardin Emeraude, Habitation Ceron and Morne Rouge) (Tab. 1). All sites in the Lezarde river harboured crayfish over a distance of 15 km from upstream to downstream. In total, in the wild, 111 individuals were captured (56 females and 55 males) with highest numbers at Site 1 of the Lezarde river (29) (the most downstream) (Tab. 1; Fig. 1). Total lengths of crayfish ranged from 4.9 cm (Manzo and Grand Galion) to 22.1 cm (Lezarde Site 1) (Tab. 1). No crayfish were caught or observed in the Oman river where specimens had been previously detected in 2014 (Tab. 1), or in the Carbet flowing only 10 meters from the hatchery's ponds (Fig. 1).
3.3 Geographic distribution of the haplotypes
A total of eight haplotypes were found in Martinique (H1–H8). The first (H1) included the majority of individuals (45), consisting of individuals from Lezarde river (24), Grand Galion (9), Mangatal hatchery (5), Petite Riviere Site 1 (5), Manzo (1) and Morne Rouge (1) and was identical to one of the haplotypes from Indonesia. The second major haplotype (H2) included 39 individuals from the Mangatal hatchery (12), Petite Lezarde (10), Lezarde river (5), Coulisses (4), Morne Rouge (3), Manzo (2), Cacaos (1), Petite Riviere Site 1 (1) and Lorrain (1). The H3 haplotype was represented by individuals from Manzo (6), Cacaos (4) and Coulisses (1) and was identical to reference haplotypes from Indonesia and Australia. The H4 haplotype contained individuals from Mangatal hatchery (4) and from Manzo (1). The H5 haplotype was only found in two individuals from Grand Galion. The H6, H7 and H8 haplotypes were rare, being represented by one individual each from Petite Riviere Site 1, Cacaos and Petite Riviere Site 1, respectively. Only three haplotypes (H1, H2 and H4) were found among the 21 crayfish sampled from the Mangatal hatchery. Haplotypes H9 and H10 contained one individual each, from the reference haplotypes from Indonesia and Malaysia (Figs. 2 and 3; Tab. 2). The mean nucleotide diversity among all haplotypes was 0.0014.
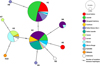 |
Fig. 2 A haplotype network using COI sequences (491 nucleotides length) of Cherax quadricarinatus collected in Martinique. A haplotype network inferred by a Median-Joining method, using 111 COI sequences of C. quadricarinatus populations from 10 different locations in Martinique (105 individuals), from Indonesia (four individuals), from Australia (one individual) and from Malaysia (one individual). The size of each circle represents the frequencies of the haplotype, with each colour showing the geographical origin of the isolates. Hatch marks indicate the numbers of mutations between haplotypes. |
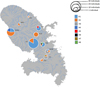 |
Fig. 3 Haplotype frequencies of Cherax quadricarinatus from 105 individuals sequenced in the 10 different locations in Martinique. Using data from Table 2 and Figure 2, diagrams are built, proportionally to the number of sequenced individuals, and each haplotype (H1–H8) was differentiated by one colour. Site names are encoded as follow: Lorrain (LOR), Morne Rouge (MRO), Mangatal (MANG), GAL (Grand Galion), Lezarde (LEZ), Petite Lezarde (PLEZ), Petite Riviere (PRIV), Manzo (MANZ), Cacaos (CAC) and Coulisses (COU). |
4 Discussion
Our results showed that the redclaw crayfish is now well established in the Martinican hydrographical networks, with 16 populations identified since its first introduction in 2004. The situation is alarming because 10 new populations were discovered since the previous survey in 2015. This number is probably an underestimation of the real situation for several reasons: (i) not all streams have yet been surveyed, (ii) capture efficiency of crayfish by traps depends on population size (Harper et al., 2018) and activity of crayfish, which can be dependent on meteorological and hydrological conditions and lunar cycles at the time of trapping (Grandjean et al., 2000).
Our results also showed that this crayfish, once introduced, can quickly colonize an entire watershed, as revealed in the Lezarde watercourse where the crayfish were sampled at four sites along 15 km of the river. This result confirms the high dispersal capabilities of this species as also reported by Nunes et al. (2017) with an average dispersal rate of 6.6 km/year. Surprisingly, the redclaw crayfish was not caught in three sites of the Carbet River adjacent to the Mangatal hatchery's ponds. This can be explained by the fact that this river has a torrential flow which would not favour the persistence of C. quadricarinatus which are found mainly in lentic or more stable and slowly flowing lotic systems (Jones, 1990).
The number of individuals caught varied among sites and ranged from 0 to 29 suggesting some populations may be present at high density, for example in the Lezarde. This is not really surprising, because C. quadricarinatus is found in Martinique freshwater ecosystems favourable for its development, suggesting a general absence of both competing species and large predators (Lim et al., 2002). The physical parameters measured in this study revealed a temperature range from 25.4 to 28.8 °C with oxygen concentrations from 2.77 to 8.06 mg/L and pH from 6.78 to 7.49 in the sites where crayfish were present (Supp. Data 1). These ranges are in accordance with the ecological preferences of C. quadricarinatus (Karplus et al., 1998; de Moor, 2002; Tropea et al., 2010). In these conditions, females could reach sexual maturity within four months and generate up to five broods per year (de Moor, 2002; Bortolini et al., 2007; Tropea et al., 2010; Arzola-Gonzalez et al., 2012; Mangatal, local farmer, pers. com.) which could explain their rapid expansion and their high density in the Lezarde river. These ecological conditions found in the rivers of Martinique seem to be excellent for the growth of this crayfish confirming by the size of trapped crayfish, up to 22 cm in total length (and 214 g) for the largest one in Lezarde river. These appear to be larger than those caught in Costa Rica, with a maximum total length of 9.22 cm (Azofeifa-Solano et al., 2017). As a result, this crayfish represents one of the biggest species present in these fresh waters, able to capture and consume large prey species and having significant ecological impacts due to their broad dietary preferences and population densities.
The introduction of C. quadricarinatus to Martinique remains poorly documented. According to the popular opinion, however, the red claw crayfish was introduced in 2004 from Cuba when a few specimens (around 20) were brought by the owner of the major crustacean hatchery on the island (Mangatal pers. com.). Results from COI sequencing from 21 individuals sampled from the Mangatal hatchery revealed three haplotypes (H1, H2 and H4) of which H1 and H2 haplotypes were represented in most of the sites where crayfish were trapped. This is therefore consistent with the Mangatal stock being the principal origin of most of the populations established in the wild in Martinique and it is not surprising because the crayfish are sold alive as a food item. They are valued as food by local people and sold at a high price (24 euros/kilo), this increases the likelihood of their introduction and eventual establishment into local ponds and watercourses enhancing the availability of this species. Two additional haplotypes were recorded in three geographically closed populations (Manzo, Coulisses, Cacaos), with at least three mutations, that were not present in the Mangatal pool. As the initial stock came from 20 individuals including both males and females, it would seem unlikely the 8 haplotypes came from this original stock. So, we cannot exclude other independent introductions of this crayfish from older farming activities. Redclaw crayfish was previously sold (before the prohibition by the law on Exotic Species (JORF n°0229 of 19th september 2020 Text n° 3)) in pet shops in Martinique until 2019. Being highly regarded by aquarists, as it often exhibits a strong blue color in aquaria, it could have been bought from local pet shops or imported and released in the wild on a number of occasions. High level of genetic variability has been also recorded for other translocated species in Europe such as Procambarus clarkii (Oficialdegui et al., 2019) and Pacifastacus leniusculus (Petrusek et al., 2017), potentially also reflecting multiple introductions.
Although no documented impacts have been reported on the ecosystem in Martinique, invasive crayfish are known to have profound impacts on freshwater communities (plants and animals) due to their opportunistic, omnivorous and predatory nature. This can affects the trophic chain at all levels, by competing with autochthonous species or predating them and perturbating the entire ecosystem functioning (Loureiro et al., 2015; Jackson et al., 2017; Lang et al., 2020). For example, several studies have shown that P. clarkii (Jackson et al., 2014; Loureiro et al., 2015; Lang et al., 2020), P. leniusculus (Jackson et al., 2014; Vaeßen and Hollert, 2015) and Faxonius virilis (formerly Orconectes virilis) (Jackson et al., 2014) can cause strong decreases in species richness of insect larvae and mollusks through predation after their introductions to a new location. Studies also suggest a decrease in the abundance and diversity of aquatic plants and animals is directly related to crayfish density (Gherardi and Acquistapace, 2007; Rodríguez-Pérez et al., 2015). Thus the presence of C. quadricarinatus could be having a significant impact on Martinique freshwater biodiversity especially where this crayfish reaches high levels of abundance such as in the Lezarde river. The rapid establishment and dispersal of this crayfish may have been facilitated by the limited number of natural crayfish predators in freshwater ecosystems in Martinique. So far, the only known effective native predators found in Martinique freshwaters seem to be eels, commonly used in biological control of crayfish in Europe (Aquiloni et al., 2010). Only Anguilla rostrata occurs in Martinique (Lim et al., 2002), but this species is now placed on the red list of species and by the IUCN as an endangered species. Because of their declining population, eels are unlikely to be able to reduce the crayfish densities and dispersal in Martinique.
Using traditional sampling methods, the current situation shows a spread of C. quadricarinatus throughout the Martinican water systems. The use of the environmental DNA (eDNA) technique in further studies could provide a more accurate measure of the distribution of C. quadricarinatus in Martinique streams. Recently, eDNA has been used to detect crayfish species, both endangered and invasive (Dougherty et al., 2016; Agersnap et al., 2017). This technique is becoming a very effective tool in the early detection of invasive species or in monitoring communities with greater efficiency than direct methods such as trapping or visual observation (Biggs et al., 2015; Hänfling et al., 2016).
In the global context of preservation of biodiversity, it seems urgent to develop strategies to prevent establishment of further new populations and investigating the impact of redclaw crayfish on the natural environment and native freshwater communities.
Supplementary Material
Supplementary data 1: Environmental parameters (Temperature, oxygen concentration and pH) measured at each site prospected during baited-trap survey, with Ponsel Mesure Odeon field tool. Access here
Acknowledgements
We thank the Direction de l'Environnement, de l'Aménagement et du Logement de Martinique and the Office de l'Eau de Martinique for their financial support and the Centre National de la Recherche Scientifique and the University of Poitiers for the provision of all the equipment necessary to carry out the study.
References
- Agersnap S, Larsen WB, Knudsen SW, Strand D, Thomsen PF, Hesselsøe M, Mortensen PB, Vrålstad T, Møller PR. 2017. Monitoring of noble, signal and narrow-clawed crayfish using environmental DNA from freshwater samples. PLoS ONE 12: 1–22. [CrossRef] [Google Scholar]
- Ahyong ST, Yeo DCJ. 2007. Feral populations of the Australian Red-Claw crayfish (Cherax quadricarinatus von Martens) in water supply catchments of Singapore. Biol Invasions 9: 943–946. [Google Scholar]
- Aquiloni L, Brusconi S, Cecchinelli E, Tricarico E, Mazza G, Paglianti A, Gherardi F. 2010. Biological control of invasive populations of crayfish: the European eel (Anguilla anguilla) as a predator of Procambarus clarkii . Biol Invasions 12: 3817–3824. [Google Scholar]
- Arzola-Gonzalez JF, Maya de la Cruz E, Verde Osuna LI, Burgueño García E, Miguel L, Campaña Flores LM, Rubio Gutiérrez Y. 2012. Crecimiento, densidad y rentabilidad del cultivo de langosta de agua dulce (Decapoda: Cherax quadricarinatus) en Sinaloa, Mexico. Ra Ximhai 8: 17–26. [CrossRef] [Google Scholar]
- Austin CM. 1996. An electrophoretic and morphological taxonomic study of the freshwater crayfish genus Cherax (Decapoda: Parastacidae) in Northern and Eastern Australia. Aust J Zool 44: 259–296. [Google Scholar]
- Azofeifa-Solano JC, Naranjo-Elizondo B, Rojas-Carranza AH, Cedeño-Fonseca M. 2017. Presence of the Australian redclaw crayfish Cherax quadricarinatus (von Martens, 1868) (Parastacidae, Astacoidea) in a freshwater system in the Caribbean drainage of Costa Rica. BioInvasions Rec 6: 351–355. [Google Scholar]
- Biggs J, Ewald N, Valentini A, Gaboriaud C, Dejean T, Griffiths RA, Foster J, Wilkinson JW, Arnell A, Brotherton P, Williams P, Dunn F. 2015. Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol Conserv 183: 19–28. [Google Scholar]
- Blackburn TM, Bellard C, Ricciardi A. 2019. Alien versus native species as drivers of recent extinctions. Front Ecol Environ 17: 203–207. [Google Scholar]
- Bláha M, Patoka J, Kozak P, Kouba A. 2016. Unrecognized diversity in New Guinean crayfish species (Decapoda, Parastacidae): the evidence from molecular data. Integr Zool 11: 457–468. [CrossRef] [PubMed] [Google Scholar]
- Bortolini L, Alvarez F, Rodriguez-Almaraz G. 2007. On the presence of the Australian redclaw crayfish, Cherax quadricarinatus, in Mexico. Biol Invasions 9: 615–620. [Google Scholar]
- Chucholl C, Mrugała A, Petrusek A. 2015. First record of an introduced population of the southern lineage of white-clawed crayfish (Austropotamobius ‘italicus’) north of the Alps. Knowl Manag Aquat Ecosyst 416: 1–8. [Google Scholar]
- de Moor I. 2002. Potential impacts of alien freshwater crayfish in South Africa. Afr J Aquat Sci 27: 125–139. [CrossRef] [Google Scholar]
- DEAL Martinique, ECOVIA, CREOCEAN. 2018. Diagnostic − Profil Environnemental de la Martinique. 139p. [Google Scholar]
- DEAL Martinique, ODE Martinique. 2016. Réalisation de pêches électriques − Caractérisation de la franchissabilité de 9 ouvrages hydrauliques et Caractérisation des stations de surveillance au titre de la Directive Cadre sur l'Eau Synthèse des 3 années de suivi 41p. [Google Scholar]
- Dougherty MM, Larson ER, Renshaw MA, Gantz CA, Egan SP, Erickson DM, Lodge DM. 2016. Environmental DNA (eDNA) detects the invasive rusty crayfish Orconectes rusticus at low abundances. J Appl Ecol 53: 722–732. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotechnol 3: 294–299. [Google Scholar]
- Gherardi F, Acquistapace P. 2007. Invasive crayfish in Europe: the impact of Procambarus clarkii on the littoral community of a Mediterranean lake. Freshw Biol 52: 1249–1259. [Google Scholar]
- Grandjean F, Cornuault B, Archambaud S, Bramard M, Otrebsky G. 2000. Life history and population biology of the white-clawed crayfish, Austropotamobius pallipes pallipes, in a brook from the Poitou-Charentes region (France). Bull Fr Pêche Piscic 356: 55–70. [CrossRef] [Google Scholar]
- Hall TA. 1999. BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symposium Series. [Google Scholar]
- Hänfling B, Handley LL, Read DS, Hahn C, Li J, Nichols P, Blackman RC, Oliver A, Winfield IJ. 2016. Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol Ecol 25: 3101–3119. [CrossRef] [PubMed] [Google Scholar]
- Harper K, Anucha P, Turnbull J, Bean C, Leaver M. 2018. Searching for a signal: environmental DNA (eDNA) for the detection of invasive signal crayfish, Pacifastacus leniusculus (Dana, 1852). Manag Biol Invasions 9: 137–148. [CrossRef] [Google Scholar]
- Hsieh C, Huang C, Pan Y. 2016. Crayfish plague Aphanomyces astaci detected in redclaw crayfish, Cherax quadricarinatus in Taiwan. J Invertebr Pathol 136: 117–123. [CrossRef] [PubMed] [Google Scholar]
- Jackson MC, Evangelista C, Zhao T, Lecerf A, Britton JR, Cucherousset J. 2017. Between-lake variation in the trophic ecology of an invasive crayfish. Freshw Biol 62: 1501–1510. [Google Scholar]
- Jackson MC, Jones T, Milligan M, Sheath D, Taylor J, Ellis A, England J, Grey J. 2014. Niche differentiation among invasive crayfish and their impacts on ecosystem structure and functioning. Freshw Biol 59: 1123–1135. [Google Scholar]
- Jaklič M, Vrezec A. 2011. The first tropical alien crayfish species in European waters: the redclaw Cherax quadricarinatus (Von Martens, 1868) (Decapoda, Parastacidae). Crustaceana 84: 651–665. [Google Scholar]
- Jones C. 1990. The biology and aquaculture potential of the tropical freshwater crayfish, Cherax quadricarinatus. Source OAI. 122 p. [Google Scholar]
- Karplus I, Zoran M, Milstein A, Harpaz S, Eran Y, Joseph D, Sagi A. 1998. Culture of the Australian red-claw crayfish Cherax quadricarinatus in Israel: III. Survival in earthen ponds under ambient winter temperatures. Aquaculture 166: 259–267. [Google Scholar]
- Kotovska G, Khrystenko D, Patoka J, Kouba A. 2016. East European crayfish stocks at risk: arrival of non-indigenous crayfish species. Knowl Manag Aquat Ecosyst 417: 37. [CrossRef] [EDP Sciences] [Google Scholar]
- Lang I, Evangelista C, Everts RM, Loot G, Cucherousset J. 2020. Stable resource polymorphism along the benthic littoral–pelagic axis in an invasive crayfish. Ecol Evol 10: 2650–2660. [CrossRef] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. 2015. POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6: 1110–1116. [Google Scholar]
- Lim P, Meunier FJ, Keith PA, Noël PY. 2002. Atlas des poissons et des crustacés d'eau douce de la Martinique. Collection patrimoines naturels. [Google Scholar]
- Lodge DM, Deines A, Gherardi F, Yeo DCJ, Arcella T, Baldridge AK, Barnes MA, Chadderton WL, Feder JL, Gantz CA, Howard GW, Jerde CL, Peters BW, Peters JA, Sargent LW, Turner CR, Wittmann ME, Zeng Y. 2012. Global introductions of crayfishes: evaluating the impact of species invasions on ecosystem services. Annu Rev Ecol Evol Syst 43: 449–472. [Google Scholar]
- Loureiro TG, Anastácio PSMG, Souty-Grosset C, Araujo PB, Almerão MP. 2015. Red swamp crayfish: biology, ecology and invasion − an overview. Nauplius 23: 1–19. [CrossRef] [Google Scholar]
- Madden T. 2002. Chapter 16: The BLAST sequence analysis tool. [Updated 2003 Aug 13]. In: McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US). Available from: http://www.ncbi.nlm.nih.gov/books/NBK21097/. [Google Scholar]
- Marufu L, Dalu T, Crispen P, Barson M, Simango R, Utete B, Nhiwatiwa T. 2018. The diet of an invasive crayfish, Cherax quadricarinatus (Von Martens, 1868), in Lake Kariba, inferred using stomach content and stable isotope analyses. BioInvasions Rec 7, 121–132. [Google Scholar]
- Mcquaid CD, Arenas F. 2009. Chapter 22-Biological invasions: insights from marine benthic communities. In: Wahl M (ed) Marine Hard Bottom Communities. Springer-Verlag, Berlin, p. 309–320. [Google Scholar]
- Morningstar CR, Daniel WM, Neilson ME, Yazaryan AK. 2020. The first occurrence of the Australian redclaw crayfish Cherax quadricarinatus (von Martens, 1868) in the contiguous United States. BioInvasions Rec 9: 120–126. [Google Scholar]
- Naqiuddin AS, Rahim KA, Long SM, Firdaus FF. 2016. The spread of the Australian redclaw crayfish (Cherax quadricarinatus von Martens, 1868) in Malaysia. J Sustain Sci Manag 11: 31–38. [Google Scholar]
- Nunes AL, Zengeya TA, Hoffman AC, Measey GJ, Weyl OLF. 2017. Distribution and establishment of the alien Australian redclaw crayfish, Cherax quadricarinatus, in South Africa and Swaziland. PeerJ 5: e3135. [CrossRef] [PubMed] [Google Scholar]
- Oficialdegui FJ, Clavero M, Sánchez MI, Green AJ, Boyero L, Michot TC, Klose K, Kawai T, Lejeusne C. 2019. Unravelling the global invasion routes of a worldwide invader, the red swamp crayfish (Procambarus clarkii). Freshw Biol 64: 1382–1400. [Google Scholar]
- Oficialdegui FJ, Sánchez MI, Clavero M. 2020. One century away from home: how the red swamp crayfish took over the world. Rev Fish Biol Fish 30: 121–135. [Google Scholar]
- Patoka J, Kalous L, Kopecký O. 2014. Risk assessment of the crayfish pet trade based on data from the Czech Republic. Biol Invasions 16: 2489–2494. [Google Scholar]
- Patoka J, Kalous L, Kopecký O. 2015. Imports of ornamental crayfish: the first decade from the Czech Republic's perspective. Knowl Manag Aquat Ecosyst 416: 4. [Google Scholar]
- Patoka J, Wardiatno Y, Kurikova P, Petrtyl M, Kalous L. 2016. Cherax quadricarinatus (von Martens) has invaded Indonesian territory west of the Wallace Line: evidences from Java. Knowl Manag Aquat Ecosyst 417: 39. [CrossRef] [Google Scholar]
- Patoka J, Wardiatno Y, Mashar A, Yonvitner, Wowor D, Jerikho R, Takdir M, Purnamasari L, Petrtýl M, Kalous L, Kouba A, Bláha M. 2018. Redclaw crayfish, Cherax quadricarinatus (von Martens, 1868), widespread throughout Indonesia. BioInvasions Rec 7: 185–189. [Google Scholar]
- Petrusek A, Filipová L, Kozubíková-Balcarová E, Grandjean F. 2017. High genetic variation of invasive signal crayfish in Europe reflects multiple introductions and secondary translocations. Freshw Sci 36: 838–850. [Google Scholar]
- Rodríguez-Pérez H, Hilaire S, Mesléard F. 2015. Temporary pond ecosystem functioning shifts mediated by the exotic red swamp crayfish (Procambarus clarkii): a mesocosm study. Hydrobiologia 767: 333–345. [Google Scholar]
- Romero X, Jimenez J. 2002. Histopathological survey of diseases and pathogens present in redclaw crayfish, Cherax quadricarinatus (Von Martens), cultured in Ecuador. J Fish Dis 25: 653–667. [Google Scholar]
- Seebens H, Blackburn TM, Dyer EE, Genovesi P, Hulme PE, Jeschke JM, Pagad S, Pyšek P, Winter M, Arianoutsou M, Bacher S, Blasius B, Brundu G, Capinha C, Celesti-Grapow L, Dawson W, Dullinger S, Fuentes N, Jäger H, Kartesz J, Kenis M, Kreft H, Kühn I, Lenzner B, Liebhold A, Mosena A, Moser D, Nishino M, Pearman D, Pergl J, Rabitsch W, Rojas-Sandoval J, Roques A, Rorke S, Rossinelli S, Roy HE, Scalera R, Schindler S, Štajerová K, Tokarska-Guzik B, Van Kleunen M, Walker K, Weigelt P, Yamanaka T, Essl F. 2017. No saturation in the accumulation of alien species worldwide. Nat Commun 8: 1–9. [Google Scholar]
- Sequencher®. 2016. Version 5.4.6 DNA sequence analysis software, Gene Codes Corporation, Ann Arbor, MI USA. [Google Scholar]
- Shull HC, Pérez-Losada M, Blair D, Sewell K, Sinclair EA, Lawler S, Ponniah M, Crandall KA. 2005. Phylogeny and biogeography of the freshwater crayfish Euastacus (Decapoda: Parastacidae) based on nuclear and mitochondrial DNA. Mol Phylogenet Evol 37: 249–263. [CrossRef] [PubMed] [Google Scholar]
- Snovsky G, Galil BS. 2011. The Australian redclaw crayfish Cherax quadricarinatus (von Martens, 1868) (Crustacea: Decapoda: Parastactidae) in the Sea of Galilee, Israel. Aquat Invasions 6: 35–37. [CrossRef] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577. [CrossRef] [PubMed] [Google Scholar]
- Tan MH, Gan HM, Gan HY, Lee YP, Croft LJ, Schultz MB, Miller AD, Austin CM. 2015. First comprehensive multi-tissue transcriptome of Cherax quadricarinatus ( Decapoda: Parastacidae) reveals unexpected diversity of endogenous cellulase. Org Divers Evol 16: 195–200. [Google Scholar]
- Tropea C, Piazza Y, López LS. 2010. Effect of long-term exposure to high temperature on survival, growth and reproductive parameters of the ‘redclaw’ crayfish Cherax quadricarinatus . Aquaculture 302: 49–56. [CrossRef] [Google Scholar]
- Uderbayev T, Patoka J, Beisembayev R, Petrtýl M, Bláha M, Kouba A. 2017. Risk assessment of pet-traded decapod crustaceans in the Republic of Kazakhstan, the leading country in Central Asia. Knowl Manag Aquat Ecosyst 418: 30. [CrossRef] [Google Scholar]
- Vaeßen S, Hollert H. 2015. Impacts of the North American signal crayfish (Pacifastacus leniusculus) on European ecosystems. Environ Sci Europe 27: 1–6. [CrossRef] [Google Scholar]
- Vodovsky N, Patoka J, Kouba A. 2017. Ecosystem of Caspian Sea threatened by pet-traded non-indigenous crayfish. Biol Invasions 19: 2207–2217. [Google Scholar]
- Weiperth A, Gál B, Kuříková P, Langrová I, Kouba A, Patoka J. 2018. Risk assessment of pet-traded decapod crustaceans in Hungary with evidence of Cherax quadricarinatus (von Martens, 1868) in the wild. North-West J Zool 15: 42–47. [Google Scholar]
Cite this article as: Baudry T, Becking T, Goût J-P, Arqué A, Gan HM, Austin CM, Delaunay C, Smith-Ravin J, Roques JAC, Grandjean F. 2020. Invasion and distribution of the redclaw crayfish, Cherax quadricarinatus, in Martinique. Knowl. Manag. Aquat. Ecosyst., 421, 50.
All Tables
Summary of the 34 sites surveyed during the study, including 20 rivers (29 sites) and 5 closed water bodies (non-functional hatchery, ornamental ponds or water retention pond).
Inventory of the 111 red claw crayfish COI sequences used to carry out the haplotype network analysis.
All Figures
 |
Fig. 1 Cartography of the sampling sites of Cherax quadricarinatus together with the presence sites of this species in Martinique Island during 2018 field campaign. Sampling was carried out on permanent streams and in closed water bodies. Cartography was prepared using QGIS 2.18 (Las Palmas) software (QGIS Development Team 2016). The shape of Martinique Island was imported from the database IGN and the streams were found in BD Carthage and BD Topo. |
| In the text | |
 |
Fig. 2 A haplotype network using COI sequences (491 nucleotides length) of Cherax quadricarinatus collected in Martinique. A haplotype network inferred by a Median-Joining method, using 111 COI sequences of C. quadricarinatus populations from 10 different locations in Martinique (105 individuals), from Indonesia (four individuals), from Australia (one individual) and from Malaysia (one individual). The size of each circle represents the frequencies of the haplotype, with each colour showing the geographical origin of the isolates. Hatch marks indicate the numbers of mutations between haplotypes. |
| In the text | |
 |
Fig. 3 Haplotype frequencies of Cherax quadricarinatus from 105 individuals sequenced in the 10 different locations in Martinique. Using data from Table 2 and Figure 2, diagrams are built, proportionally to the number of sequenced individuals, and each haplotype (H1–H8) was differentiated by one colour. Site names are encoded as follow: Lorrain (LOR), Morne Rouge (MRO), Mangatal (MANG), GAL (Grand Galion), Lezarde (LEZ), Petite Lezarde (PLEZ), Petite Riviere (PRIV), Manzo (MANZ), Cacaos (CAC) and Coulisses (COU). |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


