Fig. 2
Download original image
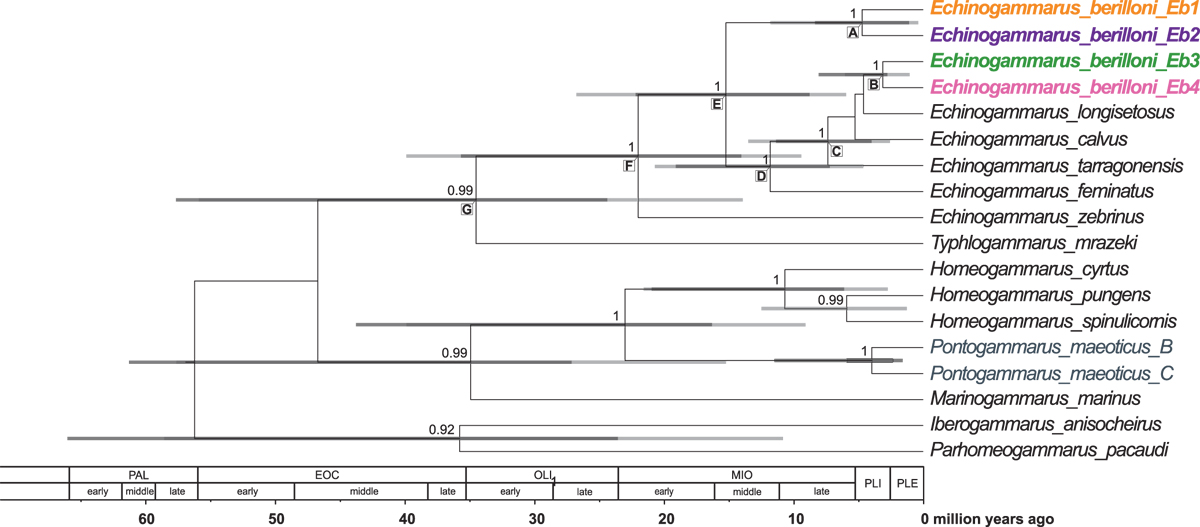
Maximum clade credibility, time-calibrated Bayesian reconstruction of phylogeny of Echinogammarus berilloni MOTUs along with the reference material. Phylogeny was inferred from sequences of the mitochondrial COI and nuclear: 28S rRNA and 18S rRNA markers. The numbers by respective nodes indicate Bayesian posterior probability values >0.9. The colours of respective MOTUs of E. berilloni correspond to those presented in Figure 1. The grey species' names as well as the highlighted nodes correspond to the sequences used for calibrating the molecular clock. Grey node bars represent 95% HPD values: light grey represent the 95% HPD values obtained using primary calibration scheme whereas dark grey bars indicate the 95% HPD values obtained using substitution rates from the literature. Further information about the divergence times of selected nodes, marked with letters, are available in Supplementary File 2.
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


