Fig. 2
Download original image
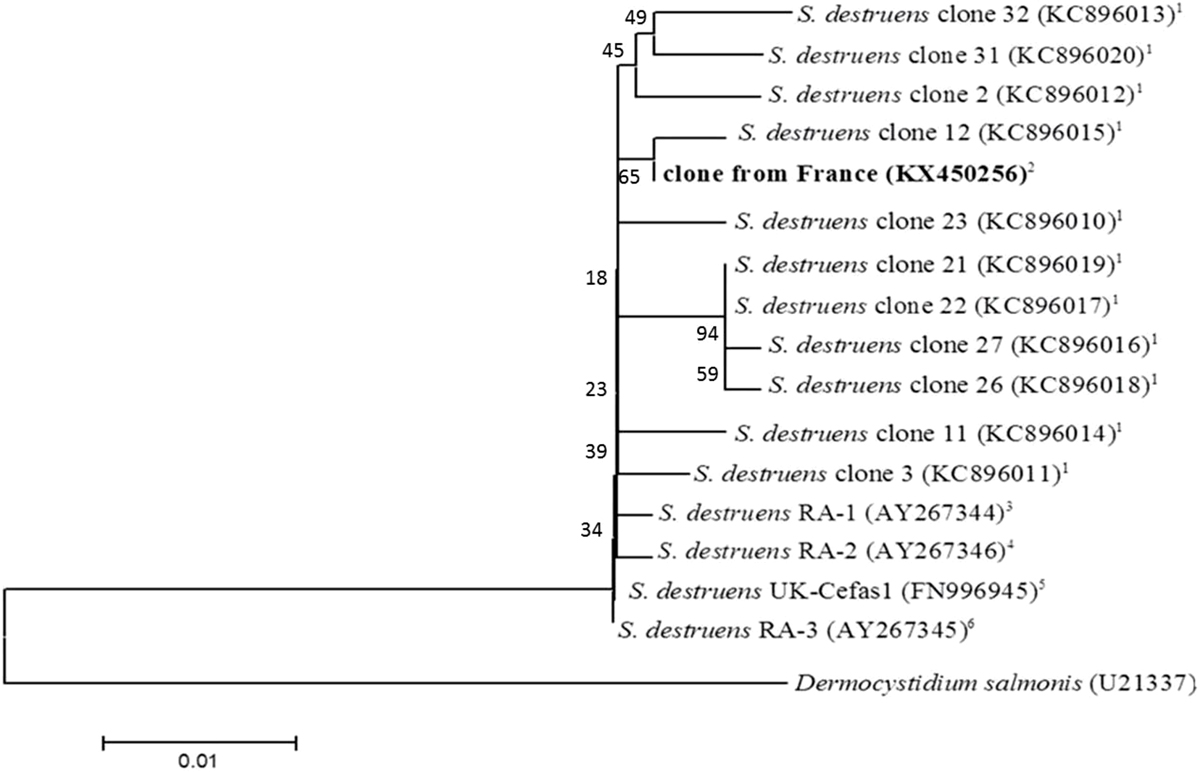
Phylogenetic tree resulting from the neighbor-joining analysis of the 18S rRNA sequences obtained from Pseudorasbora parva sampled in France and Sphaerothecum destruens sequences from GenBank, using MEGA4 software. The Dermocystidium salmonis 18S rRNA sequence was used as an outgroup. The bar represents 1% sequence divergence. The evolutionary distances were computed using the Maximum Composite Likelihood method and expressed as the number of base substitutions per site. Accession numbers are indicated in parentheses. Fish species associated with S. destruens infection, country of discovery and reference are from 1topmouth gudgeon in the Netherlands (Spikmans et al., 2013); 2topmouth gudgeon in France (this study); 3Chinook salmon in Washington, US (Arkush et al., 2003); 4Atlantic salmon in US (Arkush et al., 2003); 5sunbleak in United Kingdom (Paley et al., 2012); 6winter-run Chinook salmon in California, US (Arkush et al., 2003).
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


