| Issue |
Knowl. Manag. Aquat. Ecosyst.
Number 421, 2020
Topical issue on Crayfish
|
|
|---|---|---|
| Article Number | 40 | |
| Number of page(s) | 11 | |
| DOI | https://doi.org/10.1051/kmae/2020032 | |
| Published online | 26 October 2020 | |
Research Paper
Occurrence of two exotic decapods, Macrobrachium nipponense (de Haan, 1849) and Procambarus virginalis Lyko, 2017, in Ukrainian waters
Présence de deux décapodes exotiques, Macrobrachium nipponense (de Haan, 1849) et Procambarus virginalis Lyko, 2017, dans les eaux ukrainiennes
1
Institute of Marine Biology of the National Academy of Sciences of Ukraine, Department of water quality, 37 Pushkinska str., 65148 Odesa, Ukraine
2
V.N. Karazin Kharkiv National University, Department of Zoology and Animal Ecology, 4 Maidan Svobody, 61022 Kharkiv, Ukraine
3
Dnipro State Agrarian and Economic University, Department of Water Bioresources and Aquaculture, 25 Serhii Yefremov str., 49600 Dnipro, Ukraine
* Corresponding author: serge.utevsky@karazin.ua
Received:
9
July
2020
Accepted:
29
September
2020
New information on the occurrence of two alien decapod crustaceans, Macrobrachium nipponense and Procambarus virginalis, was summarized for Ukraine. The invasive potential of both species was estimated in the context of local environmental and socio-economic issues. Their history as non-native species is marked by the early use of alien decapods as a natural resource by local residents. Both intentional and unintentional introductions occurred for M. nipponense, while the ornamental trade was an important source of non-native P. virginalis. New records of M. nipponense and P. virginalis from open freshwater habitats in the Odesa and Kharkiv provinces suggest continuing expansion of these decapods. We conducted a molecular phylogenetic analysis of COI sequences, which confirmed morphological identifications of invasive M. nipponense. This reinforces the presumed pathway of this species, including its pathway from China to south-western Ukraine. Procambarus fallax was found to be paraphyletic in relation to P. virginalis, which was probably caused by analyzing only one mitochondrial gene, which could be impacted by the recent emergence of the latter species.
Résumé
De nouvelles informations sur la présence de deux crustacés décapodes exotiques, Macrobrachium nipponense et Procambarus virginalis, ont été compilées pour l'Ukraine. Le potentiel d'invasion des deux espÉces a été estimé dans le contexte des problèmes environnementaux et socio-économiques locaux. Leur histoire en tant qu'espèces non indigènes est marquée par l'utilisation précoce des décapodes exotiques comme ressource naturelle par les résidents locaux. Des introductions intentionnelles et non intentionnelles ont eu lieu pour M. nipponense, tandis qu'un commerce ornemental a été une source importante de P. virginalis non indigène. De nouveaux signalements de M. nipponense et de P. virginalis dans des habitats d'eau douce ouverts dans les provinces d'Odesa et de Kharkiv suggèrent une expansion continue de ces décapodes. Nous avons effectué une analyse phylogénétique moléculaire des séquences COI, qui a confirmé les identifications morphologiques de M. nipponense envahissante. Cela renforce la voie présumée de cette espèce, y compris sa trajectoire de la Chine vers le sud-ouest de l'Ukraine. On a constaté que Procambarus fallax était paraphylétique par rapport à P. virginalis, ce qui est probablement dû à l'analyse d'un seul gène mitochondrial, qui pourrait être affecté par l'émergence récente de cette dernière espèce.
Key words: Decapoda / Macrobrachium nipponense / Procambarus virginalis / Ukraine / invasive species
Mots clés : Décapode / Macrobrachium nipponense / Procambarus virginalis / Ukraine / espèce envahissante
© M.O. Son et al., Published by EDP Sciences 2020
 This is an Open Access article distributed under the terms of the Creative Commons Attribution License CC-BY-ND (https://creativecommons.org/licenses/by-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. If you remix, transform, or build upon the material, you may not distribute the modified material.
This is an Open Access article distributed under the terms of the Creative Commons Attribution License CC-BY-ND (https://creativecommons.org/licenses/by-nd/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. If you remix, transform, or build upon the material, you may not distribute the modified material.
1 Introduction
Over the past decades, expansions of five non-native decapods, Rhithropanopeus harrisii (Gould, 1841), Eriocheir sinensis Milne-Edwards, 1853, Macrobrachium nipponense (De Haan, 1849), Orconectes limosus (Rafinesque, 1815) and Procambarus virginalis Lyko, 2017 have been recorded in Ukraine's inland waters (Son et al., 2013; Novitsky and Son, 2016; Kudryashov, 2020). While invasive crabs R. harrisii and E. sinensis are common along the Azov-Black Sea coast and penetrate river basins through estuarine systems, O. limosus, M. nipponense and P. virginalis are distributed exclusively in inland waters (Son et al., 2013; Novitsky and Son, 2016; Kudryashov, 2020).
This is rather a large number of non-native species, considering that the native Ukrainian freshwater fauna includes only four decapod species, Astacus leptodactylus (Eschscholtz, 1823), Astacus astacus (Linnaeus, 1758), Astacus pachypus (Rathke, 1837) and Potamon ibericum (Bieberstein, 1808), of which the last three are not widespread and are considered to be vulnerable (Akimov, 2009; Policar et al., 2018).
Macrobrachium nipponense is a widespread Indo-Pacific species that occurs in estuarine and fresh waters (Yu and Miyake, 1972; Cai and Ng, 2002). It has been introduced to natural habitats in Bangladesh, Singapore, the Philippines, Kazakhstan, Uzbekistan, Iraq, Iran, Moldova and Ukraine (Alekhnovich and Kulesh, 2001; Cai and Shokita, 2006; De Grave and Ghane, 2006; Salman et al., 2006; Son et al., 2013). In addition, the species is cultivated in cooling reservoirs of thermal power stations in Russia, Belarus and Moldova (Alekhnovich and Kulesh, 2001).
The marmorkrebs (P. virginalis) is a triploid parthenogenetic species that was originally found in aquariums (Martin et al., 2010a, 2016). This crayfish is viewed as a recent descendant of Procambarus fallax (Hagen, 1870), which is a diploid sexually reproducing species known from Florida and Georgia, USA (Taylor et al., 1996).
The emergence of the marmorkrebs is explained by a very recent, in terms of evolution, macromutation in P. fallax that resulted in instant reproductive isolation (Vogt et al., 2015). This reproductive isolation is the basis for the assignment of the invasive marmorkrebs to its own species (Lyko, 2017). This crayfish is known as a non-native species with a high invasive potential, and it occurs throughout Europe (Martin et al., 2010b; Bohman et al., 2013; Chucholl, 2014; Vojkovská et al., 2014; Lőkkös et al., 2016; Lipták et al., 2017; Pârvulescu et al., 2017; Ercoli et al., 2019).
This research was aimed at tracing the invasion history of M. nipponense and P. virginalis in Ukraine over the last few years based on all currently available data. Both morphological and molecular characters were used to confirm the species identity and to reveal phylogenetic relationships of their populations in Ukraine.
2 Material and methods
2.1 Samples and sampling methods
In 2018–2019, decapod specimens were collected in different types of habitats in three of Ukraine's provinces representing a variety of climatic and ecological conditions of the country (Tab. 1, Figs. 1 and 2).
In a former quarry called Kotlovan Lake (Dnipropetrovsk Region), which was transparent enough to see objects of interest situated under water, samples were simply collected by hand when diving. A transect video was used to estimate population density during this collection. In waterbodies in the Akkarzhanka River Basin, a kick-net (ISO 7828) was dragged along the shore over 5 m distances. A collector moved in zigzags to cover depths from the water edge to a depth of one meter for 15 min, with 3–4 replicates. In the Great Adzhalyk Liman Basin, one specimen was sampled by hand. Similarly, in Kharkiv, crayfish samples were collected by hand in a man-made pond.
Collected specimens were immediately preserved in 96% ethanol for further morphological examination and molecular analysis. The specimens are stored in the collections of invertebrates at the Institute of Marine Biology, National Academy of Sciences of Ukraine, V. N. Karazin Kharkiv National University and the Department of Invertebrate Zoology and Hydrobiology (University of Lodz, Poland). Procambarus virginalis is kept as a living culture at the Dnipro State Agrarian and Economic University.
Samples used in our study and main characteristics of localities.
 |
Fig. 1 Habitats of exotic decapodes. A, Olexandrivka Reservoir in a small river basin; B, urban pond “Rodnyk” (Kharkiv); C, former quarry Kotlovan Lake (Dnipro); D, artificial ornamental pond (Odesa) during the renovation that caused the eradication of Procambarus virginalis. |
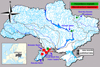 |
Fig. 2 Distribution of Procambarus virginalis and Macrobrachium nipponense in the river basins of Ukraine. See Table 1 for numbering. |
2.2 Morphological identification
Procambarus. virginalis (Fig. 3a–c) differs from the rest of alien сambarids of Europe in its specific marble coloring and very small chelipeds. Since P. virginalis belongs to the family Cambaridae, it can be reliably distinguished from all local crayfishes (Astacidae), which bear a median carina of the rostrum (Fig. 3d).
Macrobrachium nipponense (Fig. 3e–g) can be distinguished from the congeneric M. rosenbergii (De Man, 1879) by its relatively straight rostrum. The latter species has a narrow arcuate rostrum, where the tip is distinctly bent up. Macrobrachium nipponense differs from related local native palaemonid shrimps in a double row of setae along the inferior margin of the rostrum and more than ten dorsal teeth on the latter. All native species have a single row of setae along the inferior margin and no more than nine dorsal teeth of the rostrum (González-Ortegón and Cuesta, 2006). Its easily noticeable distinguishing feature is its long second pereiopods (they exceed the length of the rest of the body in larger adults) that have claws bearing numerous setae.
 |
Fig. 3 Diagnostic features of exotic decapodes. Procambarus virginalis: A, general view; B, heliped; C, rostrum without median carina. D, rostrum of Astacidae (Astacus leptodactylus) with median carina. Macrobrachium nipponense: E, general view; F, rostrum; G, claw. |
2.3 DNA extraction and PCR
The collected specimens of P. virginalis and M. nipponense were transferred to the Laboratory of Biogeography and Invertebrate Ecology, Department of Invertebrate Zoology and Hydrobiology (University of Lodz, Poland) for molecular analysis. A small piece of muscle tissues was taken for each DNA extraction. Genomic DNA was isolated using the standard phenol-chloroform method (Hillis et al., 1996). Air-dried DNA pellets were eluted in 100 µl of TE buffer, pH 8.00, stored at 4 °C until amplification, and subsequently kept at −20 °C for a long-term storage.
The standard animal DNA barcode gene region of mitochondrial cytochrome c oxidase subunit one (cox1, COI) was selected for molecular analyses (Hebert et al., 2003). It was amplified using the following primers (Folmer et al., 1994): HCO2198, 5′- TAAACTTCAGGGTGACCAAAAAATCA-3′ and LCO1490, 5′-GGTCAACAAATCATAAAGATATTGG-3′. The amplification was conducted under the following PCR conditions: 94 °C for 3 min; 5 cycles of 30 s at 94 °C, 1:30 min at 45 °C, and 1 min at 72 °C; 35 cycles of 30 s at 94 °C, 1:30 min at 51 °C, and 1 min at 72 °C; 5 min of denaturation step at 72 °C (Hou et al., 2007). PCR products (5 µl) were cleaned up using Exonuclease I (20 U/µl; EURx, Poland) and alkaline phosphatase Fast Polar-BAP (1 U/µl, EURx, Poland) treatment, following the manufacturer's guidelines. Cleaned PCR product was then sequenced by Macrogen Inc. (the Netherlands) using the same primers as at the amplification stage. The resulting sequences were manually assembled to a uniform length by removing the remaining parts of the primer regions using FinchTV v. 1.5.0 (Geospiza Inc.). These sequences were then submitted to GenBank (accession numbers: MT881672–MT881675). The length of the resulting COI sequences is 635–642 bp for P. virginalis, and 630–641 bp for M. nipponense.
2.4 Phylogenetic analysis
BLAST searches (blast.ncbi.nlm.nih.gov/Blast.cgi) were performed to check for the amplification of contaminants. The sequences of the COI gene region were aligned using MAFFT ver.7 online (Katoh et al., 2017) with the FFT-NS-i strategy selected by the “Auto” option. The COI sequences were also checked for mistaken stop codons at the amino acid level using Molecular Evolutionary Genetics Analysis (MEGA) program version X (Kumar et al., 2018).
To perform our phylogenetic analysis, previously published GenBank nucleotide sequences (Salman et al., 2006; Feng et al., 2008; Wowor et al., 2009; Martin et al., 2010a, 2010b; Filipová et al., 2011; Liu and Cui, 2011; Aoki et al., 2013; Bohman et al., 2013; Quan et al., 2014; Vojkovská et al., 2014; Vogt et al., 2015; Usio et al., 2017 Cui et al., 2018; Zheng et al., 2019) were combined with our molecular data. Localities and GenBank accession numbers of sequences used in the present study are listed in Supplementary Table S1.
The phylogeny was reconstructed using Maximum Likelihood (Strimmer and von Haeseler, 1996) with the interior branch tests of 1000 bootstrap replicates (Sitnikova et al., 1995) using MEGA X. Analyses were based on the HKY model with a discrete Gamma distribution [+G, α = 0.0500] for the P. virginalis phylogeny and Tamura 3-parameter model (Tamura, 1992) with a discrete Gamma distribution [+G, α = 0.5572] for the M. nipponense phylogeny, as they were the best substitution models computed in MEGA X using the Bayesian information criterion. The tree was drawn to scale with branch lengths measured in the number of substitutions per site.
In addition, the number of base differences per site (p-distances) between sequences and their standard errors were calculated. All positions containing gaps and missing data were eliminated. Haplotypes were determined using DnaSP version 6.12.03 (Rozas et al., 2017). There were a total of 614 positions for P. virginalis and 551 positions for M. nipponense in the final dataset.
3 Results
3.1 Field research
Several sequential attempts were made to find P. virginalis based on previous records and anecdotal evidences. This approach revealed some fluctuations in the occurrence of this species. In a previous study (Novitsky and Son, 2016), P. virginalis was first recorded for Lake Kotlovan on 23 October 2015. However, according to Alexander Trushlyakov's personal communication, in the summer of 2014, large numbers of P. virginalis already occurred in this locality. Later, in November 2015, P. virginalis was not found, which could be explained by a low temperature (+6 °C) at that time. In the autumn of 2018 (11 October 2018), the species was again recorded in that locality in the course of night sampling. In the shallow waters of the lake, we counted up to 2–3 individuals per 100 square meters of a diving transect. Later, on 16 November 2018, 34 adult individuals and 2 juveniles of P. virginalis were caught by means of night sampling.
One of the first recorded localities of P. virginalis − a cascade of ornamental ponds in Odesa, where three live individuals were observed in June 2015 (Novitsky and Son, 2016) − was also surveyed. Shortly after that discovery, those reservoirs underwent multi-stage cleaning and renovation (Fig. 1d). During the winter-spring period of 2017–2018, a consistent complete drying of all ponds was carried out. In the course of those activities, large animals such as fish and turtles, including invasive red-eared sliders Trachemys scripta (Schoepff, 1792), were placed in aquariums and released back later. Conversely, soft sediment accumulated at the bottom along with benthic invertebrates was removed from these water bodies. As a result, we can confidently assert that P. virginalis was eradicated in that locality.
One individual of P. virginalis was found in a man-made pond in Kharkiv on 10 October 2019. There were no previous information on its occurrence in Kharkiv and neighboring areas.
Outside the Dniester Basin, we found M. nipponense in the basin of the small river Akkarzhanka, which flows into the estuarine Sukhyi Liman. Several specimens of the species were sampled on 23 July 2018. Later, a density of up to 15 individuals per square meter was estimated in August 2018. In all samplings, young individuals were found.
Five other ponds of the Sukhyi Liman basin were examined, and the shrimp was also found in the Oleksandrivka Reservoir on 2 September 2018 with a density of 3–4 individuals (adults, including females with eggs) per square meter.
Only one specimen was sampled in a reservoir of the Great Adzhalyk Liman Basin on 13 March 2020. This locality and the Oleksandrivka Reservoir represent a rare type of habitat − the upper estuary separated from the rest of the estuary by dams and turned into a freshwater body which is still highly mineralized.
3.2 Molecular studies
The molecular phylogenetic analysis revealed that all examined specimens of P. virginalis both from the Dnipropetrovsk and Kharkiv regions were identical in all 614 base pairs to COI GenBank sequences from Germany, Japan, Italy, and Sweden (LC228303, KJ690261, KT074364, KF033123, HM35810, HM358011, JF438007) (Martin et al., 2010a, 2010b; Filipová et al., 2011; Bohman et al., 2013; Vojkovská et al., 2014; Usio et al., 2017) (see Supplem. Tab. S2). Remarkably, COI sequences for the Ukrainian samples are also identical to the holotype of P. virginalis (accession number: KT074364), which was described from Germany (Vogt et al., 2015; Lyko, 2017). The phylogenetic relationships are shown on the tree in Figure 4. See Table 2 for between and within group nucleotide divergence estimation.
The phylogenetic analysis of COI sequences of M. nipponense did not reveal novel haplotypes. The whole alignment consisted of 551 base pairs with 29 variable sites. The haplotypes and their affiliation to all sequences obtained are shown in Supplementary Table S3.
Our sequences were identical to those of M. nipponense from Gaoming Disrict, Foshan, Guangdong Province, China (MK412772) and M. nipponense obtained during a survey of biodiversity in the Yangtze River, China, especially from the largest freshwater lakes (JN874532) (Feng et al., 2008; Zheng et al., 2019). The Maximum Likelihood tree based on COI sequences is shown in Figure 5. The between and within group nucleotide divergence estimation is shown in Table 3.
 |
Fig. 4 Maximum Likelihood phylogenetic tree of Procambarus virginalis and related species based on COI gene sequences (highest log likelihood-1285.73). Samples from the current research are highlighted in bold, grey box showing sequences with the same haplotype. Bootstrap values are shown for clades. See Table S1 for details of the analyzed specimens and their GenBank accession numbers. |
Estimates of average evolutionary divergence over sequence pairs of the P. virginalis dataset. The number of base differences per site (based on p-distances) between groups and within each group are shown.
 |
Fig. 5 Maximum Likelihood phylogenetic tree of Macrobrachium nipponense based on COI gene sequences (highest log likelihood-2143.56). Samples from the current research are highlighted in bold, grey box showing sequences with the same haplotype. See Table S1 for the details of the analyzed specimens and their GenBank accession numbers. |
Estimates of average evolutionary divergence over sequence pairs of the M. nipponense dataset. The number of base differences per site (based on p-distances) between groups and within each group are shown.
4 Discussion
4.1 Identification and phylogenetic relationships
Morphological characters and molecular data suggest assigning the specimens examined to two decapod species, M. nipponense and P. virginalis. The sequences of the latter were identical to GenBank samples from Germany, Sweden, Italy and Japan in its COI genes, thus sharing the same haplotype. This can be explained by a recent invasion, which has not provided sufficient time for the divergence of their mitochondrial genes. Macrobrachium nipponense was found to have low genetic diversity as well; Ukrainian samples share the same haplotype with two individuals from China. The shallow phylogenetic structure revealed in both the species is in agreement with the founder effect hypothesis, which is commonly used to explain the recent evolutionary history of a number of invasive species (Estoup et al., 2016).
Procambarus virginalis is also known under its former name Procambarus fallax forma virginalis (Martin et al., 2010a, 2010b). In 2017, the systematic rank of this taxon was elevated to the level of species (Lyko, 2017). It occurs in Germany and elsewhere, while the native range of its parental species P. fallax is restricted to southern Georgia and Florida, USA (Hobbs, 1989). In our study, we compared the COI gene region of our samples to the complete genome of P. virginalis, which was published alongside with its formal taxonomic description (Lyko, 2017), and sequences of P. fallax from Florida (Martin et al., 2010b), which is usually considered as its native range. The phylogenetic analysis revealed that the Ukrainian marmorkrebs shared the same haplotype with the type specimen of P. virginalis, whereas sequences of P. fallax from the USA formed separate clades. Remarkably, the genetic distance observed between these two species is low and just slightly exceeds the within group mean p-distances observed in them. Procambarus fallax, which is a parental species in relation to P. virginalis, was found to be paraphyletic. Although species are usually expected to be monophyletic groups, this topology can be viewed as a consequence of analyzing only one mtDNA gene (Coyne and Orr, 2004). The paraphyly can be explained by incomplete lineage sorting due to the recency of the emergence of P. virginalis, which still retains its relationships with certain ancestral populations of the parental species. Currently, the overall genetic structure of P. virginalis cannot be estimated with certainty due to the lack of reliable distinguishing between P. virginalis and P. fallax sequences in GenBank. One or very few nearly identical COI haplotypes of P. virginalis are expected to exist.
The phylogenetic relationships of the Ukrainian M. nipponense corroborate the known invasive history of the species (see below), confirming the Chinese origin of the populations studied.
4.2 Invasion history
Previous sources have provided contradictory information on the origin of the Dniester population of M. nipponense. Both a Chinese (Alekhnovich and Kulesh, 2001) and Japanese (Stepanok, 2014) origin of the shrimp has been supposed. Based on available information, we traced all stages of this invasion (Fig. 6).
In the 1960s, this species was accidentally introduced (along with young herbivorous fish) from the Yangtze River (China) to some former Soviet regions. As a result of this, stable populations were reported from water bodies of Uzbekistan and Kazakhstan (Alekhnovich and Kulesh, 2001) and from the cooling pond of the Elektrogorsk thermal power station “GRES-3” on outskirts of Moscow (Ivanov and Starobogatov, 1974). The latter population was a source for subsequent successful intentional introductions to the cooling ponds of the thermal power stations “Berezovskaya GRES” (Belarus) and “Moldavskaya GRES” (Moldova) (Vladimirov et al., 1989; Khmeleva et al., 1997; V.A. Kulesh, personal communication).
Before the 2000s, M. nipponense had not been reported from the Dniester Basin, except the Kuchurhan Liman (an estuary on the border of Ukraine and Moldova associated with the Dniester Delta and impacted by the thermal power station “Moldavskaya GRES”). Records from the Dniester Delta started to occur in the 2000s (Son et al., 2013). In 2013–2016, the next expansion of the range was revealed. Macrobrachium nipponense was recorded in two separate areas of the Dniester Liman (Stepanok, 2014), which is the estuary of the Dniester River; in the Moldovan Dniester near Tiraspol (Filipenko, 2014); and in fishponds in the Ukrainian Dniester Delta (Shekk and Astafurov, 2017), which became a key event for its further expansion. These fishponds are associated with the largest local fish farms in the area, where fishes are bred for subsequent stocking in other fishponds in the Lower Dniester, Lower Danube and neighboring small river basins. Our findings of this species in small river basins in 2018–2020 are most likely explained by this vector.
Moreover, since 2017, freshwater shrimps were observed in waterbodies near the town of Sarata, Odesa Region. Based on photographs from local Internet sources (e.g., https://bessarabiainform.com/2018/09/vozle-saraty-rybaki-nahodyat-gigantskih-krevetok-fotofakt/), they can be provisionally identified as M. nipponense. In addition, the shrimp from the fishponds in the Dniester Delta were repeatedly caught in the abovementioned farms for keeping in small decorative ponds in private gardens in Odesa (personal communications from local fishermen). Thus, its range may be wider than this research suggests.
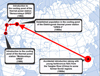 |
Fig. 6 Invasion history of Ukrainian populations of Macrobrachium nipponense. A, China; B, Russia; C, Belarus; D, Moldova and Ukraine. |
4.3 Pathways and risks of secondary distribution
Both P. virginalis and M. nipponense are sold in ornamental animal markets. The ornamental vector of introduction for P. virginalis has been discussed many times and looks to be highly likely for Ukrainian records. The marbled crayfish is popular in all large pet markets of Ukraine and in online trade, where they are cheaper than other exotic cambarid species.
Urban water reservoirs are a very common target for introduction of ornamental species by aquarium hobbyists as a result of market saturation, which promotes illegal releases of commercially unprofitable and unwanted juveniles to urban waters (Novitsky and Son, 2016). With regard to P. virginalis, this is true because both its parthenogenetic reproduction mode and rapid growth (Martin et al., 2010a) make its population difficult to control even in aquaria.
In recent years P. virginalis has become one of the most popular ornamental decapods, while Macrobrachium species which were the most popular decapods kept by local aquarists 20 years ago, are now sold much less frequently than other shrimp species. However, the combination of the intentional introduction of M. nipponense and its random distribution with stocking individuals poses a high risk of invasion into new river basins. Apparently, climate changes are making the Ponto-Caspian region increasingly suitable for Macrobrachium species. For example, these species have been recorded in the Iranian Caspian Sea basin and in the Don Delta (De Grave and Ghane, 2006; Shokhin, 2018). The invasion to Sarata's waterbodies can be a starting point for the further penetration of Macrobrachium into local freshwater ecosystems, as it is connected with the Danube Delta through the Sarata River channel, Lake Sasyk and the Danube-Sasyk Canal.
With regard to P. virginalis, the localities in Dnipro and Kharkiv promote the possibility of its expansion into the Dnieper and Don basins. Both of these localities are situated in areas close to the Dnieper-Donbas Canal, which connects the Siversky Donets sub-basin with the Dnieper and promotes exchange of alien species (Semenchenko et al., 2015; Son et al., 2014).
4.4 Harmful effects of invasive decapods
The crayfish plague, caused by the fungus-like pathogen Aphanomyces astaci Schikora, 1906, has been identified as key threat to European crayfish from non-indigenous crayfish species (Holdich et al., 2009). Recently the presence of the plague pathogen has been confirmed in a wild population of the marbled crayfish in Germany (Keller et al., 2014). Aphanomyces astaci was long considered a specialist pathogen of crayfish; however, freshwater shrimps and crabs have now also been found to be vectors of this parasite (Svoboda et al., 2014; Schrimpf et al., 2014; Mrugała et al., 2019). Recent studies have revealed that this pathogen is present in the Dnieper (Ukraine) and Dniester (Moldova) basins (Panteleit et al., 2018; Ungureanu et al., 2020).
Nutritional studies of M. nipponense in eutrophic freshwater wetlands of the Caspian coast, which are very close in terms of environmental conditions to southern Ukraine, show that this species can be a significant additional predator of invertebrates. Its diet also includes other components of food webs: detritus, algae, macrophytes, etc. (Mirzajani et al., 2020).
4.5 Economic use
Giant shrimps of the genus Macrobrachium play a key role in modern freshwater aquaculture. Beginning in Soviet times, experimental cultivation of M. nipponense and M. rosenbergii was established in some natural and warm-water reservoirs (Suprunovich and Makarov, 1990). Macrobrachium rosenbergii is cultivated in reservoirs on the Crimean Peninsula, but no information is known about its escape from aquaculture or acclimatization in Ukraine's natural ecosystems. The invasive populations of both P. virginalis and M. nipponense instantly began to be used by local people as a food resource. According to Alexander Trushlyakov's personal communication, in Dnipro, P. virginalis was caught by local people in large numbers in 2014 and sold at small local markets for food. Similarly, in fish ponds of the Dniester Delta, immediately after the invasion of M. nipponense, pond owners began to catch them specifically to sell as food or for stocking (according communications from local fishermen).
Supplementary Material
Supplementary Table S1: List of localities and GenBank accession numbers of sequences used in the present study.
Supplementary Table S2: Variable sites in the alignment of COI sequences of the P. fallax complex with 5 different haplotypes of 17 sequences used in this study.
Supplementary Table S3: Variable sites in the alignment of COI sequences of M. nipponense with 9 different haplotypes of 20 sequences used in this study.
Access hereAcknowledgements
The molecular analysis was performed during H.M.'s fellowship for molecular taxonomy at the Department of Invertebrate Zoology and Hydrobiology, University of Lodz, Poland. This research was funded in part by the statutory funds at the University of Lodz. We are warmly grateful to Prof. Michał Grabowski and all lab members for helping, teaching and encouraging H.M. during the fellowship. We gratefully acknowledge divers Alexander Trushlyakov and Anatoly Artemenko for their field assistance. We are warmly grateful to Prof. Vladimir F. Kulesh (Belarusian State Pedagogical University) for his valuable consultations. Our special thanks go to Dr. Michael Tessler (American Museum of Natural History, USA) for linguistic improvement of the manuscript.
References
- Akimov IA. (ed.) 2009. Red book of Ukraine, animals, Kyiv: Globalconsulting, 600 p. [Google Scholar]
- Alekhnovich AV, Kulesh VF. 2001. Variation in the parameters of the life cycle in prawns of the genus Macrobrachium Bate (Crustacea, Palaemonidae). Russ J Ecol 32: 454–459. [CrossRef] [Google Scholar]
- Aoki M, Hamasaki K, Yamada M. 2013. Identification of three Macrobrachium species from the Kii Peninsula, Japan using PCR-RFLP. Jap J Limnol 74: 85–91 (In Japanese). [CrossRef] [Google Scholar]
- Bohman P, Edsman L, Martin P, Scholtz G. 2013. The first Marmorkrebs (Decapoda: Astacida: Cambaridae) in Scandinavia. BioInvasions Rec 2: 227–232. [Google Scholar]
- Cai Y, Ng PKL. 2002. The freshwater palaemonid prawns of Myanmar (Crustacea: Decapoda: Caridea). Hydrobiologia 487: 59–83. [Google Scholar]
- Cai Y, Shokita S. 2006. Report on a collection of freshwater shrimps (Crustacea: Decapoda: Caridea) from the Philippines, with descriptions of four new species. Raffles Bull Zool 54: 245–270. [Google Scholar]
- Chucholl C. 2014. Predicting the risk of introduction and establishment of an exotic aquarium animal in Europe: insights from one decade of Marmorkrebs (Crustacea, Astacida, Cambaridae) releases. Manag Biol Invasions 5: 309–318. [CrossRef] [Google Scholar]
- Coyne JA, Orr HA. 2004. Speciation. Sunderland, MA: Sinauer Associates, 545 p. [Google Scholar]
- Cui F, Yu Y, Bao F, Wang S, Xiao MS. 2018. Genetic diversity analysis of the oriental river prawn (Macrobrachium nipponense) in Huaihe River. Mitochondrial DNA Part A 29: 737–744. [CrossRef] [Google Scholar]
- De Grave S, Ghane A. 2006. The establishment of the oriental river prawn, Macrobrachium nipponense (de Haan, 1849) in Anzali Lagoon, Iran. Aquat Invasions 1: 204–208. [Google Scholar]
- Ercoli F, Kaldre K, Paaver T, Gross R. 2019. First record of an established marbled crayfish Procambarus virginalis (Lyko, 2017) population in Estonia. Bioinvasions Rec 8: 675–683. [Google Scholar]
- Estoup A, Ravigné V, Hufbauer R, Vitalis R, Gautier M, Facon B. 2016. Is there a genetic paradox of biological invasion? Annu Rev Ecol Evol S 47: 51–72. [CrossRef] [Google Scholar]
- Feng JB, Sun YN, Cheng X, Li JL. 2008. Sequence analysis of mitochondrial COI gene of Macrobrachium nipponense from the five largest freshwater lakes in China. J Fish China 32: 517–525 (In Chinese). [Google Scholar]
- Filipenko SI. 2014. The appearance of the freshwater eastern shrimp Macrobrachium nipponense (De Haan, 1849) in Dniester River. In: Sustainable Use and Protection of Animal World Diversity: International Symposium Dedicated to 75th Anniversary of Professor Andrei Munteanu, Tipografia Academiei de Ştiinţe a Moldovei, Chișinău, pp. 206–207 (In Russian). [Google Scholar]
- Filipová L, Grandjean F, Chucholl C, Soes DM, Petrusek A. 2011. Identification of exotic North American crayfish in Europe by DNA barcoding. Knowl Manag Aquat Ecosyst 401: 1–14. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3: 294–299. [PubMed] [Google Scholar]
- González-Ortegón E, Cuesta J. 2006. An illustrated key to species of Palaemon and Palaemonetes (Crustacea: Decapoda: Caridea) from European waters, including the alien species Palaemon macrodactylus . J Mar Biol Assoc U K 86: 93–102. [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, Dewaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci 270: 313–321. [Google Scholar]
- Hillis DM, Moritz C, Mable BK. 1996. Molecular systematic, 2nd ed. Sunderland, Massachusetts: Sinauer Associates, 655 p. [Google Scholar]
- Hobbs Jr HH. 1989. An illustrated checklist of the American crayfishes (Decapoda, Astacidae, Cambaridae, Parastacidae). Smithson Contr Zool 480: 1–236. [CrossRef] [Google Scholar]
- Holdich DM, Reynolds JD, Souty-Grosset C, Sibley PJ. 2009. A review of the ever increasing threat to European crayfish from non-indigenous crayfish species. Knowl Manag Aquat Ecosyst 11: 394–395. [Google Scholar]
- Hou ZG, Fu JH, Li SQ. 2007. A molecular phylogeny of the genus Gammarus (Crustacea: Amphipoda) based on mitochondrial and nuclear gene sequences. Mol Phylogenet Evol 45: 596–611. [Google Scholar]
- Ivanov BG, Starobogatov YI. 1974. Subtropical freshwater prawns Macrobrachium nipponense (Palaemonidae) in the waters of the Moscow region. Ekologiya 6: 83–85 (In Russian). [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20: 1160–1166. [Google Scholar]
- Keller NS, Pfeiffer M, Roessink I, Schulz R, Schrimpf A. 2014. First evidence of crayfish plague agent in populations of the marbled crayfish (Procambarus fallax forma virginalis). Knowl Manag Aquat Ecosyst 414: 15. [CrossRef] [EDP Sciences] [Google Scholar]
- Khmeleva NN, Kulesh VF, Alekhnovich AV, Giginyak YG. 1997. Ecology of freshwater prawns, Belaruskaya Navuka, Minsk, 254 p. (In Russian) [Google Scholar]
- Kudryashov SS. 2020. Materials on fish protected on the national level and new invasive crustacean species in the Danube and the Danube lakes. In: Monitoring and Protection of Biodiversity in Ukraine: Animals, Druk Art, Kyiv-Chernivtsi, pp. 128–129. [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35: 1547–1549. [Google Scholar]
- Lipták B, Mojžišová M, Gruľa D, Christophoryová J, Jablonski D, Bláha P, Petrusek A, Kouba A. 2017. Slovak section of the Danube has its well-established breeding ground of marbled crayfish Procambarus fallax f. virginalis . Knowl Manag Aquat Ecosyst 418: 1–5. [Google Scholar]
- Liu Y, Cui Z. 2011. Complete mitochondrial genome of the Chinese spiny lobster Panulirus stimpsoni (Crustacea: Decapoda): genome characterization and phylogenetic considerations. Mol Biol Rep 38: 403–410. [PubMed] [Google Scholar]
- Lőkkös A, Müller T, Kovács K, Várkonyi L, Specziár A, Martin P. 2016. The alien, parthenogenetic marbled crayfish (Decapoda: Cambaridae) is entering Kis-Balaton (Hungary), one of Europe's most important wetland biotopes. Knowl Manag Aquat Ecosyst 417: 1–9. [Google Scholar]
- Lyko F. 2017. The marbled crayfish (Decapoda: Cambaridae) represents an independent new species. Zootaxa, 4363: 544–552. [CrossRef] [PubMed] [Google Scholar]
- Martin P, Dorn N, Kawai T, van der Heiden C, Scholtz G. 2010a. The enigmatic Marmorkrebs (marbled crayfish) is the parthenogenetic form of Procambarus fallax (Hagen, 1870). Contrib Zool 79: 107–118. [CrossRef] [Google Scholar]
- Martin P, Shen H, Füller G., Scholtz G. 2010b. The first record of the parthenogenetic Marmorkrebs (Decapoda, Astacida, Cambaridae) in the wild in Saxony (Germany) raises the question of its actual threat to European freshwater ecosystems. Aquat Invasions 5: 397–403. [Google Scholar]
- Martin P, Thonagel S, Scholtz G. 2016. The parthenogenetic Marmorkrebs (Malacostraca: Decapoda: Cambaridae) is a triploid organism. J Zool Syst Evol Res 54: 13–21. [Google Scholar]
- Mrugała A, Buřič M, Petrusek M, Kouba A. 2019. May atyid shrimps act as potential vectors of crayfish plague? NeoBiota 51: 65–80. [CrossRef] [Google Scholar]
- Mirzajani A, Ghane A, Bagheri S. Abbasi K, Sayadrahim M, Salahi M, Lavajoo F. 2020. Diet Survey and Trophic Position of Macrobrachium nipponense in the Food Web of Anzali Wetland. Wetlands, 1–11. [Google Scholar]
- Novitsky RA, Son MO. 2016. The first records of Marmorkrebs Procambarus fallax (Hagen, 1870) f. virginalis (Crustacea, Decapoda, Cambaridae) in Ukraine. Ecol Montenegrina 5: 44–46. [Google Scholar]
- Panteleit J, Keller NS, Diéguez-Uribeondo J, Makkonen J, Martín-Torrijos L, Patrulea V, Pîrvu M, Preda C, Schrimpf A, Pârvulescu L. 2018. Hidden sites in the distribution of the crayfish plague pathogen Aphanomyces astaci in Eastern Europe: Relicts of genetic groups from older outbreaks? J Invertebr Pathol 157: 117–124. [CrossRef] [PubMed] [Google Scholar]
- Pârvulescu L, Togor A, Lele S-F, Scheu S, Șinca D, Panteleit J. 2017. First established population of marbled crayfish Procambarus fallax (Hagen, 1870) f. virginalis (Decapoda, Cambaridae) in Romania. BioInvasions Rec 6: 357–362. [Google Scholar]
- Policar T, Bondarenko V, Bezusyj O, Stejskal V, Kristan J, Malinovskyi O, Imentai A, Blecha M, Pylypenko Y. 2018. Crayfish in Central and Southern Ukraine with special focus on populations of indigenous crayfish Astacus pachypus (Rathke, 1837) and their conservation needs. Aquat Conserv 28: 6–16. [Google Scholar]
- Quan AS, Pease KM, Breinholt JW, Wayne RK. 2014. Origins of the invasive red swamp crayfish (Procambarus clarkii) in the Santa Monica mountains. Aquat Invasions 9: 211–219. [Google Scholar]
- Rozas J, Ferrer-Mata A, Snchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Snchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34: 3299–3302. [CrossRef] [PubMed] [Google Scholar]
- Salman SD, Page TJ, Naser MD, Yasser AG. 2006. The invasion of Macrobrachium nipponense (De Haan, 1849) (Caridea: Palaemonidae) into the Southern Iraqi marshes. Aquat Invasions 1: 109–115. [Google Scholar]
- Semenchenko VP, Son MO, Novitsky RA, Kvatch YV, Panov VE. 2015. Alien macroinvertebrates and fish in the Dnieper River Basin. Russ J Biol Invasions 6: 51–64. [CrossRef] [Google Scholar]
- Schrimpf A, Schmidt T, Schulz R. 2014. Invasive Chinese mitten crab (Eriocheir sinensis) transmits crayfish plague pathogen (Aphanomyces astaci). Aquat Invas 9: 203–209. [CrossRef] [Google Scholar]
- Shekk PV, Astafurov YO. 2017. The influence of diet composition and growing conditions on the cannibalism in eastern freshwater shrimp Macrobrachium nipponense (de Haan, 1849). Rybogospodarska Nauka Ukrainy 40: 49–59 (In Russian). [CrossRef] [Google Scholar]
- Shokhin IV. 2018. The first find of shrimp of the genus Macrobrachium Bate, 1868 (Crustacea: Decapoda: Palaemonidae) in the Don River. Science in the South of Russia 14: 109–112 (In Russian). [CrossRef] [Google Scholar]
- Sitnikova T, Rzhetsky A, Nei M. 1995. Interior-branch and bootstrap tests of phylogenetic trees. Mol Biol Evol 12: 319–333. [PubMed] [Google Scholar]
- Son MO, Novitsky RA, Dyadichko VG. 2013. Recent state and mechanisms of invasions of exotic decapods in Ukrainian rivers. Vestn Zool 47: 59–64. [CrossRef] [Google Scholar]
- Son MO, Prokin AA, Dubov PG, Konopacka A, Grabowski M, MacNeil C, Panov VE. 2020. Caspian invaders vs. Ponto-Caspian locals - range expansion of invasive macroinvertebrates from the Volga Basin results in high biological pollution of the Lower Don River. Manag Biol Invasions 11: 178–200. [CrossRef] [Google Scholar]
- Stepanok NA. 2014. Oriental river prawn of the genus Macrobrachium in the lower reach of the Dniester River. Hydrobiol J 50: 110–113. [CrossRef] [Google Scholar]
- Strimmer K, von Haeseler A. 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol Biol Evol 13: 964–971. [Google Scholar]
- Suprunovich AV, Makarov YN. 1990. Food invertebrates: mussels, oysters, scallops, cryfishes, shrimps. Kiev: Naukova dumka, 264 p. [Google Scholar]
- Svoboda J, Strand DA, Vrålstad T, Grandjean F, Edsman L, Kozák P, Kouba A, Fristad RF, Koca SB, Petrusek A. 2014. The crayfish plague pathogen can infect freshwater-inhabiting crabs. Freshw Biol 59: 918–929. [Google Scholar]
- Tamura K. (1992). Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol 9: 678–687. [PubMed] [Google Scholar]
- Taylor CA, Warren ML, Fitzpatrick JF, Hobbs HH, Jezerinac RF, Pflieger WL, Robinson HW. 1996. Conservation status of crayfishes of the United States and Canada. Fisheries 21: 25–38. [CrossRef] [Google Scholar]
- Ungureanu E, Mojžišová M, Tangerman M, Ion MC, Pârvulescu L, Petrusek A. 2020. The spatial distribution of Aphanomyces astaci genotypes across Europe: introducing the first data from Ukraine. Freshwater Crayfish 25: 77–87. [Google Scholar]
- Usio N, Azuma N, Sasaki S, Oka T, Inoue M. 2017. New record of Marmorkrebs from western Japan and its potential threats to freshwater ecosystems. Cancer 26: 5–11. [Google Scholar]
- Vladimirov MZ, Toderash IK, Chorik FP. 1989. Oriental freshwater prawn (Macrobrachium nipponense De Haan) − the new element of the Kuchurgan reservoir hydrofauna. Izvestiya AN MSSR Ser Biol 1: 77–78 (In Russian). [Google Scholar]
- Vogt G, Falckenhayn C, Schrimpf A, Schmid K, Hanna K, Panteleit J, Helm M, Schulz R, Lyko F. 2015. The marbled crayfish as a paradigm for saltational speciation by autopolyploidy and parthenogenesis in animals. Biology Open 4: 1583–1594. [Google Scholar]
- Vojkovská R, Horká I, Tricarico E, Ďuris Z. 2014. New record of the parthenogenetic marbled crayfish Procambarus fallax f. virginalis from Italy. Crustaceana 87: 1386–1392. [Google Scholar]
- Wowor D, Muthu V, Meier R, Balke M, Cai Y, Ng PK. 2009. Evolution of life history traits in Asian freshwater prawns of the genus Macrobrachium (Crustacea: Decapoda: Palaemonidae) based on multilocus molecular phylogenetic analysis. Mol Phyl Evol 52: 340–350. [CrossRef] [Google Scholar]
- Yu H-P, Miyake S. 1972. Five species of the genus Macrobrachium (Crustacea, Decapoda, Palaemonidae) from Taiwan. Ohmu 3: 45–55. [Google Scholar]
- Zheng XZ, Chen WJ, Guo ZL. 2019. The genus Macrobrachium (Crustacea, Caridea, Palaemonidae) with the description of a new species from the Zaomu Mountain Forest Park, Guangdong Province, China. ZooKeys 866: 65–83. [CrossRef] [PubMed] [Google Scholar]
Cite this article as: Son MO, Morhun H, Novitskyi RO, Sidorovskyi S, Kulyk M, Utevsky S. 2020. Occurrence of two exotic decapods, Macrobrachium nipponense (de Haan, 1849) and Procambarus virginalis Lyko, 2017, in Ukrainian waters. Knowl. Manag. Aquat. Ecosyst., 421, 40.
All Tables
Estimates of average evolutionary divergence over sequence pairs of the P. virginalis dataset. The number of base differences per site (based on p-distances) between groups and within each group are shown.
Estimates of average evolutionary divergence over sequence pairs of the M. nipponense dataset. The number of base differences per site (based on p-distances) between groups and within each group are shown.
All Figures
 |
Fig. 1 Habitats of exotic decapodes. A, Olexandrivka Reservoir in a small river basin; B, urban pond “Rodnyk” (Kharkiv); C, former quarry Kotlovan Lake (Dnipro); D, artificial ornamental pond (Odesa) during the renovation that caused the eradication of Procambarus virginalis. |
| In the text | |
 |
Fig. 2 Distribution of Procambarus virginalis and Macrobrachium nipponense in the river basins of Ukraine. See Table 1 for numbering. |
| In the text | |
 |
Fig. 3 Diagnostic features of exotic decapodes. Procambarus virginalis: A, general view; B, heliped; C, rostrum without median carina. D, rostrum of Astacidae (Astacus leptodactylus) with median carina. Macrobrachium nipponense: E, general view; F, rostrum; G, claw. |
| In the text | |
 |
Fig. 4 Maximum Likelihood phylogenetic tree of Procambarus virginalis and related species based on COI gene sequences (highest log likelihood-1285.73). Samples from the current research are highlighted in bold, grey box showing sequences with the same haplotype. Bootstrap values are shown for clades. See Table S1 for details of the analyzed specimens and their GenBank accession numbers. |
| In the text | |
 |
Fig. 5 Maximum Likelihood phylogenetic tree of Macrobrachium nipponense based on COI gene sequences (highest log likelihood-2143.56). Samples from the current research are highlighted in bold, grey box showing sequences with the same haplotype. See Table S1 for the details of the analyzed specimens and their GenBank accession numbers. |
| In the text | |
 |
Fig. 6 Invasion history of Ukrainian populations of Macrobrachium nipponense. A, China; B, Russia; C, Belarus; D, Moldova and Ukraine. |
| In the text | |
Current usage metrics show cumulative count of Article Views (full-text article views including HTML views, PDF and ePub downloads, according to the available data) and Abstracts Views on Vision4Press platform.
Data correspond to usage on the plateform after 2015. The current usage metrics is available 48-96 hours after online publication and is updated daily on week days.
Initial download of the metrics may take a while.


